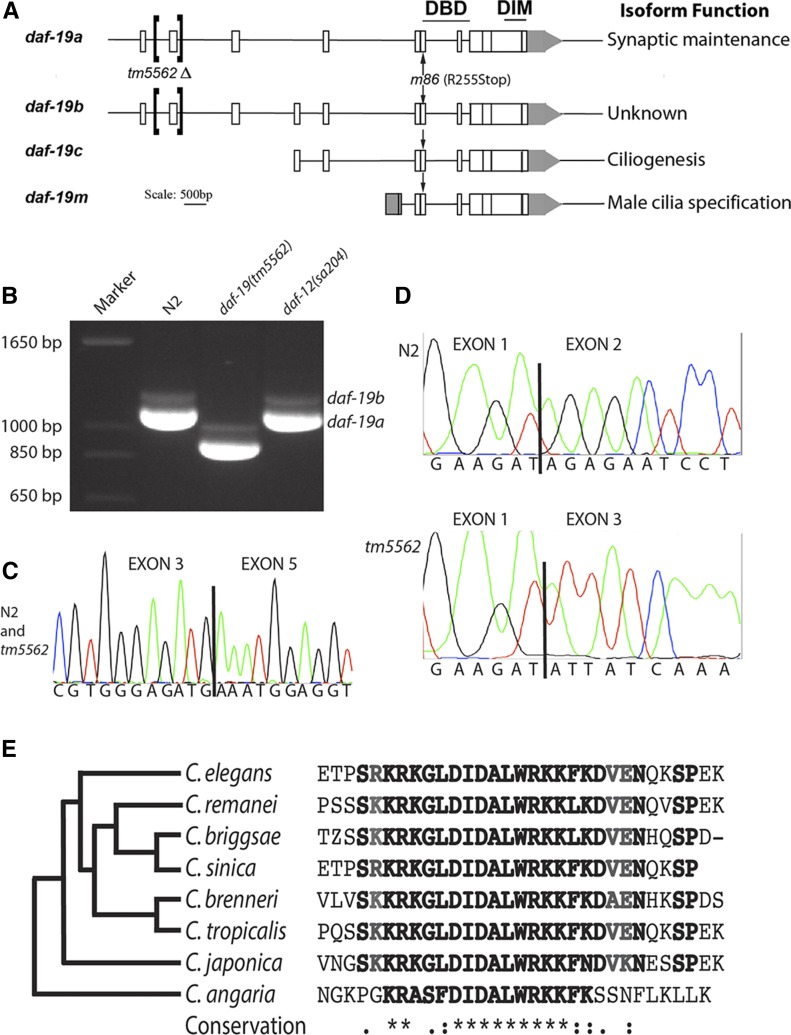
Figure 1.
daf-19 structure and characterization of daf-19(tm5562) RNA products. (A) Three promoters produce four related daf-19 RNA products, as shown. Conserved DNA-binding (DBD) and dimerization (DIM) domains are indicated above encoding exons; locations of a new deletion allele tm5562 and the null allele m86 are noted with brackets and arrows, respectively. (B) Reverse transcription (RT)-PCR using primers to exons 1 and 10 reveal a highly abundant 1123-bp daf-19a transcript from worms wild-type for daf-19, a 934-bp transcript from tm5562 worms, and a much less abundant daf-19b product that is 75-bp larger. (C) Sequencing trace from the smaller RT-PCR product shows splicing of exon 3–5 as in daf-19a. (D) Sequencing traces of exon 1 splicing in daf-19a cDNA products from wild-type N2 (top) and daf-19(tm5562) worms (below). (E) Translated sequences from exon 2 of daf-19 orthologs aligned using T-Coffee (Di Tommaso et al. 2011) and clustered (left) according to the phylogeny of Slos et al. (2017). Sequence identifiers: C. elegans, gi|71988112; C. remanei, gb|EFP05251.1; C. briggsae, emb|CAP22350.4; C. sinica, scaffold 899; C. brenneri, gb|EGT33833.1; C. tropicalis, scaffold 563.1; C. japonica, contig 15741.5; and C. angaria, contig 57337_2.