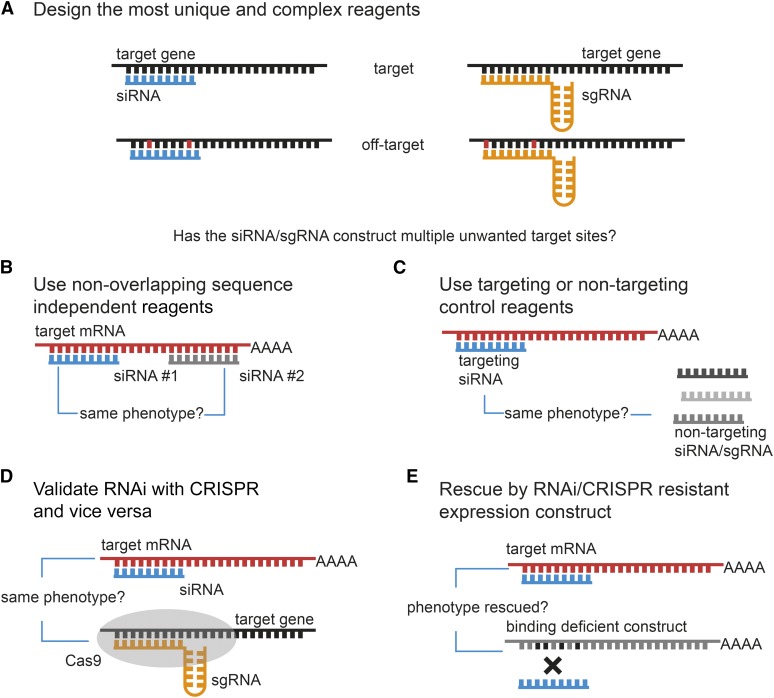
Figure 6.
Strategies for minimizing false-positive results by off-target effects. Recent research has shown that genetic perturbations by RNAi and CRISPR are not 100% precise. Phenotypic effects resulting from reagents targeting unwanted sides in the genome are termed off-target effects (OTEs). Many strategies have been developed to minimize the risk of reporting phenotypes from off-target effects. (A) One measure to avoid OTEs is by designing reagents using specialized software that carefully assesses whether reagents possess multiple possible target sides in the targeted genome. (B) Comparing phenotypes of multiple sequence independent reagents is the most widely used method for ensuring target specificity and is state of the art in all functional RNAi and CRISPR/Cas experiments. (C) Unwanted side effects resulting from cellular or organismal reactions toward the reagent injection or transfection can be controlled via the use of nontargeting or nonsense targeting reagents. (D) Controls that require follow-up experiments include the validation of RNAi knockdown phenotypes using CRISPR/Cas-driven gene knockout and vice versa. (E) A rescue of the phenotype by an RNAi or sgRNA binding deficient overexpression construct can further increase confidence that the observed phenotype results from perturbing the gene of interest.