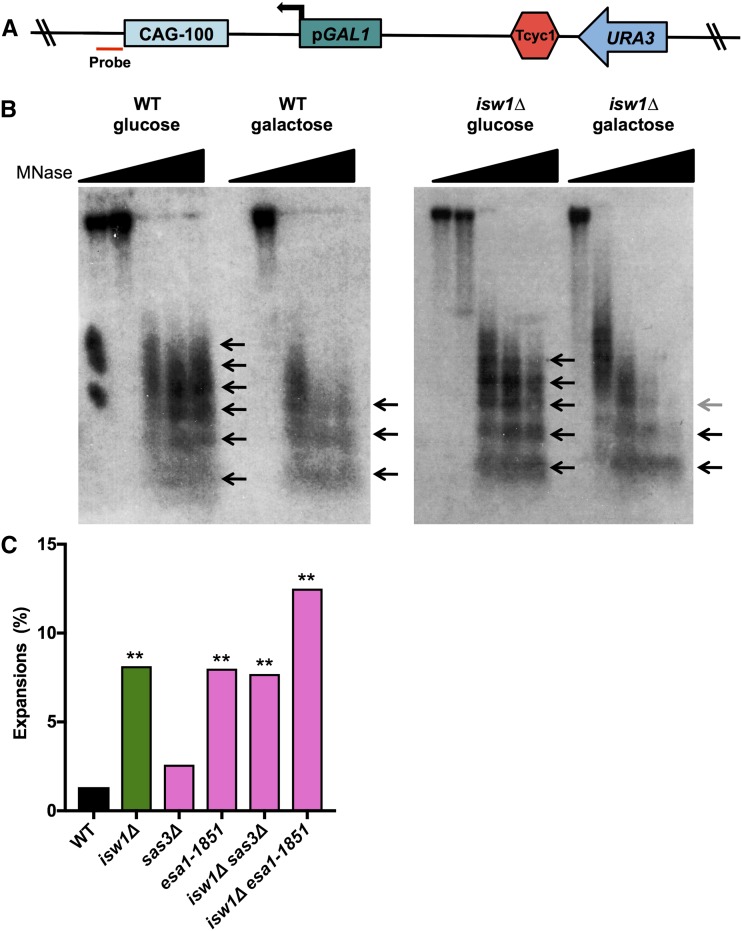
Figure 7.
Transcription reduces nucleosome occupancy over a CAG tract and isw1∆ alters the nucleosome array. (A) CAG-100 pGAL1 URA3-YAC construct showing location of the probe used in a Southern blot, 14-bp upstream of the CAG repeat (red). (B) Micrococcal nuclease (MNase) assay of wild-type (WT) and isw1∆ strains grown in noninducing (glucose) or inducing (galactose) conditions. The wedge indicates increasing MNase levels from 0 to 15 units; the 0-unit lane is missing in the WT galactose condition. The arrows indicate MNase-protected regions; an array of three to six distinguishable positioned nucleosomes is visible. Less-digested or less-positioned arrays appear as smears. (C) The frequency of CAG-85 expansions was measured in WT and isw1∆ strains with deletion of SAS3 or a catalytic mutation of ESA1 histone acetyltransferase (HAT) genes. Expansion frequencies were tested for significant deviation from WT using Fisher’s exact test; ** P < 0.01.