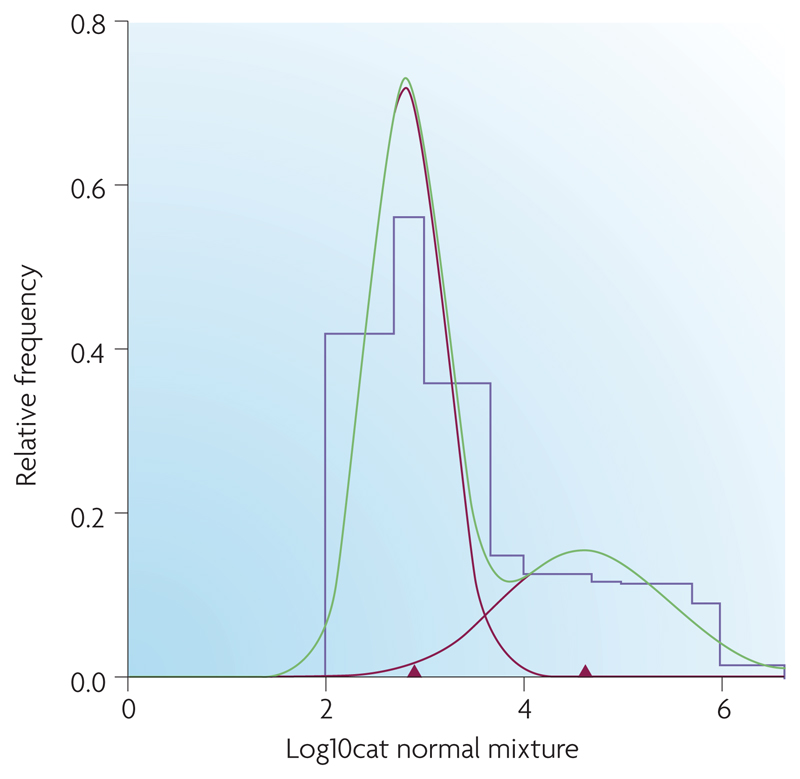
Figure 3.
Results of mixture distribution analysis on a histogram of E. coli O157 counts (log transformed) gathered from faecal pats collected during a cross-sectional survey of Scottish store and finishing cattle (2002-2004)13. This data was analyzed using MIX (Ichthus Data Systems, Hamilton, Ontario, Canada), a program that analyzes histograms as mixtures of statistical distributions. Parameters of the mixture distribution (mean, standard deviation, proportion) were calculated as maximum likelihood estimates (MLE) using a combination of the Expectation-Maximization (EM) algorithm and Newton-type methods. Final models were fitted assuming two normal components. This figure shows the original histogram of E. coli O157 counts (purple line) fitted with 2 component normal distributions (red lines). The green line is their sum, the mixture distribution, which matches the histogram as closely as possible. The red triangles mark the mean E. coli O157 count within each component distribution. The point estimate of threshold for a super- shedder was chosen as the point of overlap of the two unscaled distributions, approximately 3135 C.F.U. g-1; 95% CI for threshold estimate, 1658 and 10395. Note that this does not match the value which might be inferred from this figure, where the components are scaled relative to the sizes of the observed populations. Reproduced with permission from REF 13 © 2007American society of Microbiology.