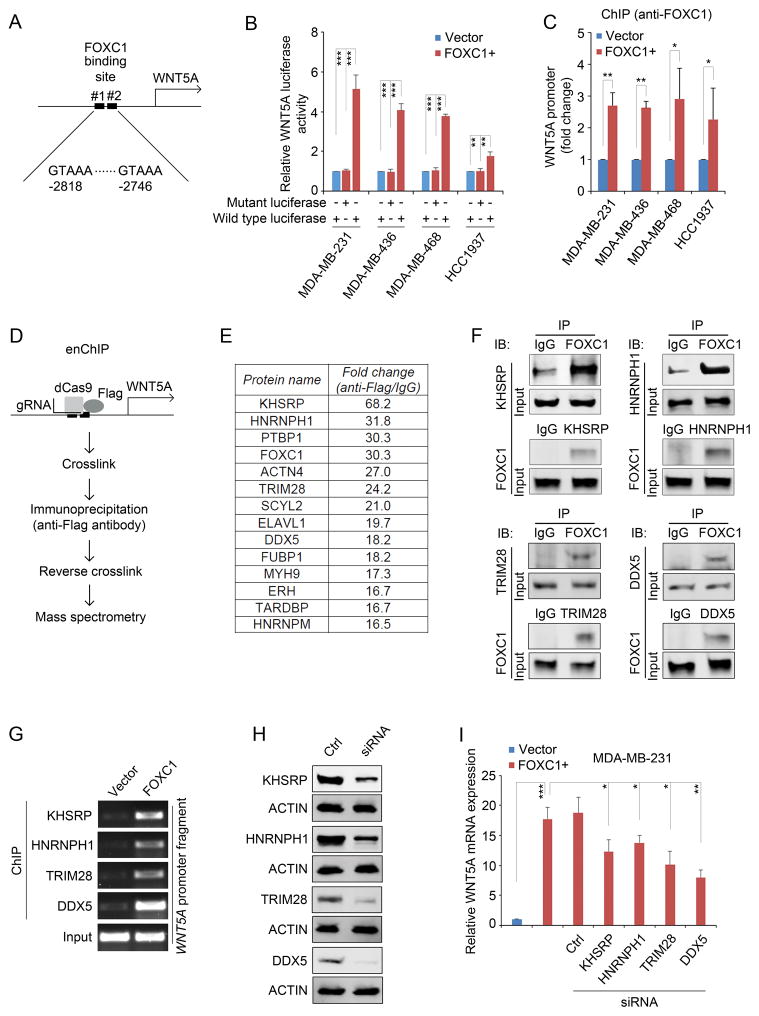
Figure 2.
WNT5A promoter is activated by a FOXC1 transactivator complex. A, diagrammatic illustration of putative FOXC1 binding sites in the WNT5A promoter. B, luciferase assays of cells transfected with the pGL4-WNT5A promoter luciferase reporter constructs containing wild-type or mutant FOXC1 binding sites. pGL4-WNT5A promoter plasmids was constructed by cloning wild-type (GTAAA) or mutant (ACCGC) 177 base pairs (−2868 to −2692, from the translation start site) of WNT5A promoter fragment into pGL4 luciferase reporter vector (Promega). The β-Galactosidase expression vector was from Promega. Cells seeded in 12-well plates were transfected with 500ng luciferase plasmids and 500ng expression plasmids using Lipofectamine 2000 (Invitrogen). Two hundred ng of β-galactosidase expression plasmid was used as an internal control. After 48h, cells were harvested and 20μl extracts were analyzed using the Luciferase Reporter Assay System (Promega) according to the manufacturer’s instructions. Luciferase activity was measured using a luminometer (Glomax multi detection system, Promega). The bar graph indicates mean ± SD, n = 3. **, p < 0.01, ***, p < 0.001. C, ChIP assays using an anti-FOXC1 antibody to pull down the WNT5A promoter-protein complex. ChIP assays were performed using the EZ-ChIP Chromatin Immunoprecipitation Kit (EMD Millipore) according to the manufacturer’s instructions. Anti-FOXC1 (sc-21394, Santa Cruz) antibody-immunoprecipitated DNA was analyzed by real-time PCR. The primers were: WNT5A-forward: 5′-AGACTGTAAAATGCCCACAGGT-3′, WNT5A-reverse: 5′-TCAAAGCTCCCCTTGGGAC-3′. The bar graph indicates mean ± SD, n = 3. *, p < 0.05, **, p < 0.01. D, diagrammatic illustration of the enChIP assay. E, the top enriched proteins that bind to the WNT5A promoter together with FOXC1. enChIP was performed as described previously.17,36 Briefly, cells were transfected with 3×FLAG-dCas9 (#51240, Addgene) and pSIR-neo-WNT5A (WNT5A gBlocks were sub-cloned into pSIR-neo (#51128, Addgene)). Guide RNA (gRNA) sequences for WNT5A: #1: 5′-CCTGGGGGCGATTTGCCGGG-3′; #2: 5′-GAAGTGGTCAAGGTTTACAG-3′; #3: 5′-TAAGGCCGCACACGCCCTGG-3′. Cells were crosslinked and chromatins were immunoprecipitated with an anti-FLAG antibody (F1804, Sigma-Aldrich). After reverse crosslinking, proteins were analyzed by liquid chromatography tandem mass spectrometry (LC-MS/MS) essentially as described.37 Briefly, following in-gel protein digestion, tryptic peptides were separated on a 50 cm EASY-Spray C18 column, and analyzed by an LTQ Orbitrap Elite mass spectrometer in the data-dependent acquisition mode. MS data were searched against the Uniprot Human database (released on 01/22/2016) with MaxQuant (v1.5.5.1).38 Protein quantification was performed using spectral counting.39 F, co-immunoprecipitation (Co-IP) assays to confirm the binding between FOXC1 and the factors identified in the enChIP assays. Co-IP was performed as described previously.9 Briefly, the whole cell lysates were extracted using IP buffer (50mM Tris-HCl (pH7.4), 150mM NaCl, 2mM EDTA-2Na, 1% NP40, 10% Glycerol) and immunoprecipitated with antibody-bound agarose beads (Thermo). Protein complexes were analyzed by western blotting analysis. The antibodies used in Co-IP assay were the same as the ones used in the western blotting. G, ChIP assays using different antibodies in control or FOXC1-overexpressing MDA-MB-231 cells. Immunoprecipitated WNT5A promoter fragments were subject to PCR assays. Primers used were: WNT5A-forward: 5′-AGACTGTAAAATGCCCACAG GT-3′, WNT5A-reverse: 5′-TCAAAGCTCCCCTTGGGAC-3′. H, western blotting analysis of the proteins in control siRNA- or protein-specific siRNA-transfected MDA-MB-231 cells. ACTIN was used as an internal control. KHSRP, HNRNPH1, TRIM28, and DDX5 siRNAs were from Santa Cruz. The primary antibodies used were: KHSRP (1:1000, #13398, Cell Signaling Technology), HNRNPH1 (1:500, sc-10042, Santa Cruz), TRIM28 (1:1000, #4123, Cell Signaling Technology), and DDX5 (1:1000, #9877, Cell Signaling Technology). I, real-time PCR analysis of WNT5A mRNA expression in MDA-MB-231 cells transfected with different siRNAs. Primers used were: WNT5A-forward: 5′-CCCTCGCCATGAAGAAGTCCA-3′, WNT5A-reverse: 5′-CATACCTAGCGACCACCAAGA -3′. The bar graph indicates mean ± SD, n = 3. *, p < 0.05, **, p < 0.01, ***, p < 0.001.