Figure 6.
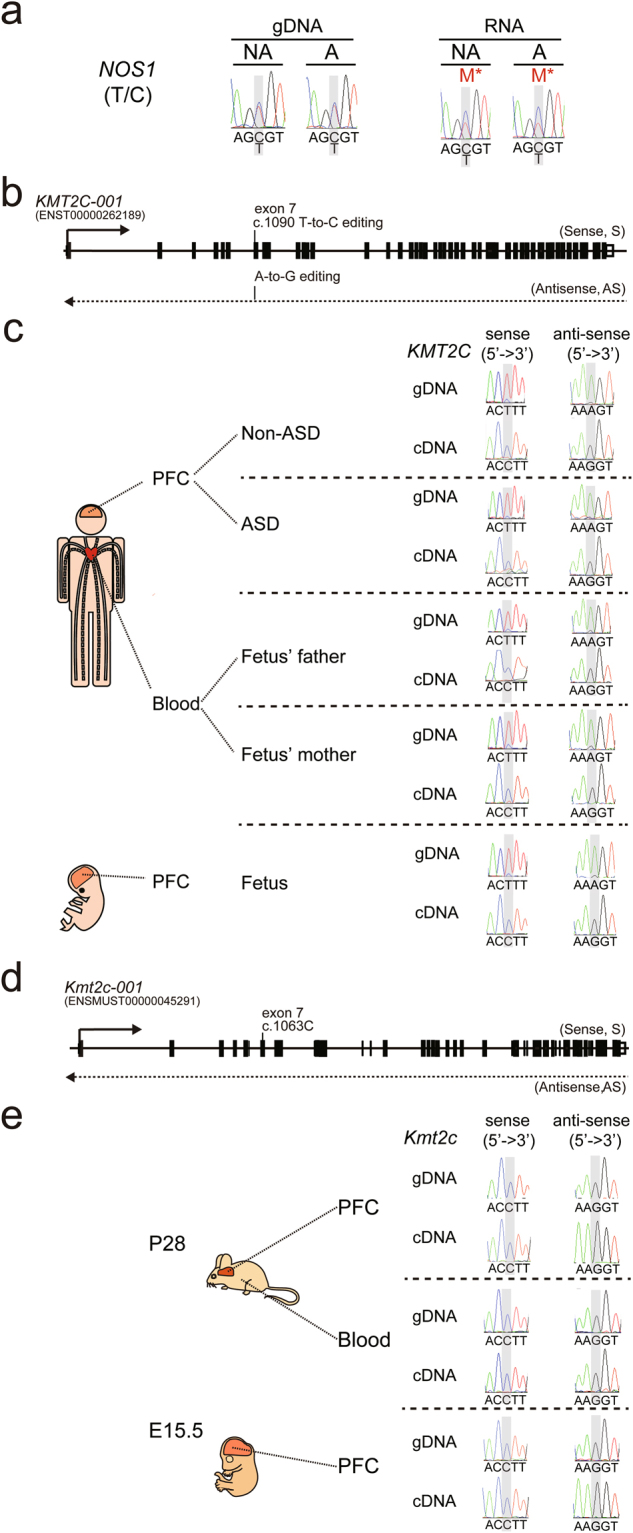
Identification of noncanonical imprinting in the PFC of the offspring without and with ASD and editing of KMT2C transcript in human PFC. (a) Sanger sequencing was used to analyze noncanonical imprinting of NOS1 in the PFC of the offspring without and with ASD. “M*” stands for maternally-biased expression. (b) Schematic diagram of the genomic locus of human KMT2C. Arrows indicate the direction of transcription. (c) Sanger sequencing was performed to analyze RNA editing for KMT2C in the PFC of the offspring without and with ASD, fetal PFC, and blood from parents of the fetus. (d) Schematic diagram of the genomic locus of mouse Kmt2c. Arrows indicate the direction of transcription. (e) Sanger sequencing was performed to analyze RNA editing for Kmt2c in PFC and blood from postnatal day 28 (P28) mice and PFC from embryonic day 15.5 (E15.5) mice. SNP information is shown as (paternal allele/maternal allele).
