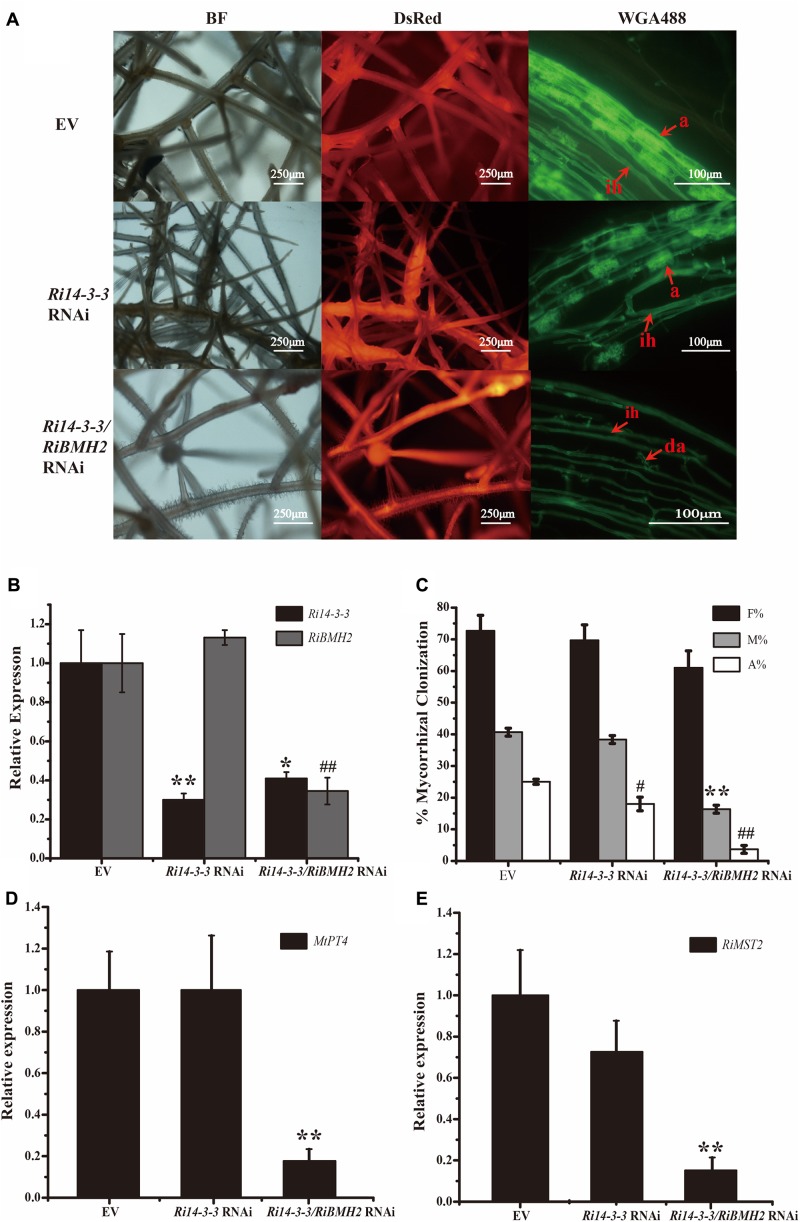
FIGURE 5.
Mycorrhizal symbiotic phenotypes of Host-Induced Gene Silencing of Ri14-3-3 and RiBMH2. (A) Hairy root transformation of M. truncatula with empty vector (EV), Ri14-3-3 RNAi vector, or Ri14-3-3/BMH2 RNAi vector. Transgenic hairy roots were infected by AM fungi and the mycorrhizal phenotypes were observed with fluorescence microscope. a, mature arbuscules; ad, arbuscule degradation; ih, internal hyphae. (B) Transcript abundance change of Ri14-3-3 and RiBMH2 in transgenic hairy roots as measured by qRT-PCR using RiActin gene as the reference gene. (C) Mycorrhization level was analyzed by WGA 488 staining of hairy roots at 30 dpi with R. irregularis. F%, frequency of colonization; M%, intensity of mycorrhiza; A%, arbuscule abundance. (D) Expression levels of MtPT4 in control (EV) and RNAi lines were determined by real-time RT-PCR. The M. truncatula MtTEF gene was used as the reference gene. (E) Transcript accumulation of RiMST2 in control (EV) and RNAi mycorrhizal roots measured by real-time RT-PCR. The R. irregularis RiActin gene was used as endogenous control. Three technical replicates were analyzed. Asterisks indicate statistically significant differences from respective control lines. Error bars indicate the means of three biological replicates with SD values. Data shown are averages ± SD; n = 3. (#, ∗p < 0.05, ##, ∗∗p < 0.01).