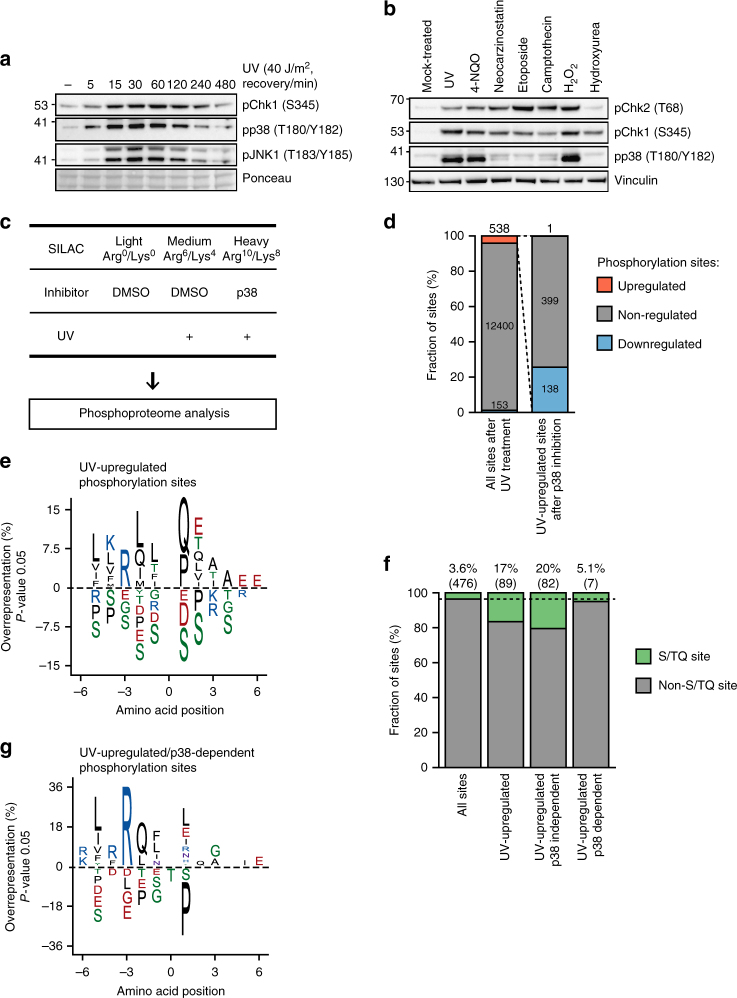
Fig. 1.
Phosphoproteomics reveals a broad scope of p38 signaling after UV light. a U2OS cells were treated with UV light (40 J/m2) and left to recover for the indicated times. Total cell lysates were resolved on SDS-PAGE and activation of p38, JNK1, and Chk1 was monitored with phospho-specific antibodies. b U2OS cells were left untreated or treated with UV light (40 J/m2, 1 h recovery), 4-nitroquinoline 1-oxide (4-NQO, 20 µM, 1 h), neocarzinostatin (400 µg/ml, 1 h), etoposide (10 µM, 1 h), camptothecin (10 µM, 1 h), H2O2 (2 mM, 1 h), and hydroxyurea (2 mM, 1 h). Total cell lysates were resolved on SDS-PAGE and blotted with the indicated antibodies. c Schematic representation of the strategy used to identify UV-light-induced, p38-dependent phosphorylation sites. SILAC-labeled U2OS cells were mock-treated (Light), irradiated with UV light (40 J/m2, 1 h recovery) (Medium), or pretreated with the p38 inhibitor (SB203580, 10 µM, 1 h) or transfected with p38 siRNA, and irradiated with UV light (40 J/m2, 1 h recovery) (Heavy). Phosphorylated peptides were enriched using TiO2 and peptide samples were analyzed by LC-MS/MS. d The bar graph shows the number of significantly up-, non-, and downregulated UV-light-induced phosphorylation sites after p38 inhibition identified from two replicate experiments (p-value < 0.01, moderated t-test). Significantly regulated phosphorylation sites were calculated as shown in Supplementary Figure 1f, g. e Sequence motif analysis of 538 UV-light-induced phosphorylation sites. The iceLogo plot shows frequency of six amino acids flanking phosphorylated residue. The frequencies of amino acids surrounding phosphorylated residues in UV-light-induced phosphorylation sites was compared with frequencies in all quantified phosphorylation sites. A significant overrepresentation of phosphorylation sites conforming to the ATM/ATR/DNA-PKcs motif (S/TQ) is observed among UV-light-induced sites. f The bar graph shows the absolute number and percentage of S/TQ sites among all quantified phosphorylation sites, UV-light-upregulated sites, UV-light-upregulated, p38-independent sites, and UV-light-upregulated, p38-dependent sites. g Sequence motif analysis of 138 UV-light-induced, p38-dependent phosphorylation sites. The analysis was done as described in Fig. 1e