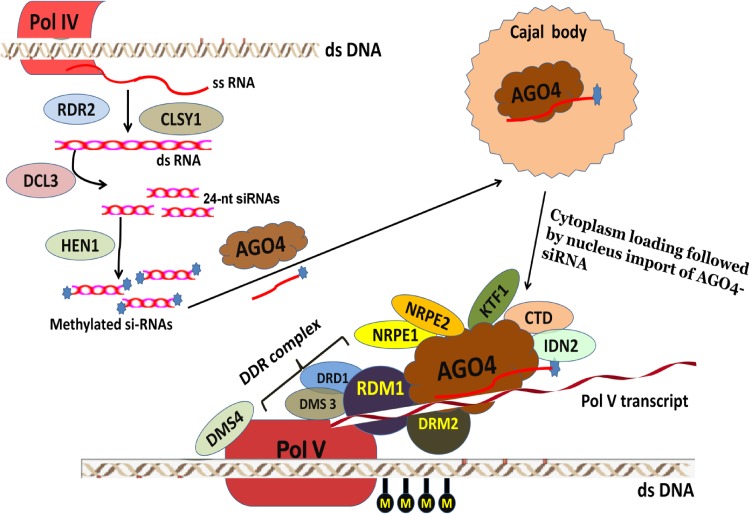
Fig. 2.
Schematic illustration of RNA-directed DNA methylation. RNA-directed DNA methylation is initiated by RNA polymerases IV (Pol IV). The single stranded RNA transcript (ss RNA) transcribed by Pol IV is then copied into double stranded RNA (ds RNA) with the help of RNA-DEPENDENT RNA POLYMERASE 2 (RDR2). CLASSY 1 (CLSY1) also termed as CHR38, is a putative chromatin remodeller or helicase that helps in recruiting Pol IV to chromatin or aid in ssRNA transcript processing. DICER-LIKE 3 (DCL3) process the dsRNA into 24-nucleotide small interfering RNA (siRNA) duplexes which are later methylated by HUA-ENCHANCER 1 (HEN1) at their 3′ ends. In the presence of ARGONAUTE 4 (AGO4), ds siRNA dissociates to form ssRNA which associates with AGO4 to form an RNA-induced silencing complex (RISC). AGO4 localizes to Cajal bodies, that seems to be necessary for wild type levels of RdDM33. Independently of siRNA synthesis, Pol V transcription is assisted by the DDR complex, comprising of DEFECTIVE IN RNA-DIRECTED DNA METHYLATION (DRD1), DEFECTIVE IN MERISTEM SILENCING 3 (DMS3), REQUIRED FOR DNA METHYLATION (RDM1) and DMS4. AGO4 binds Pol V transcripts through pairing with the siRNA and is become stable by AGO4 interaction with the NUCLEAR RNA POLYMERASE E1 (NRPE1), NRPE2, carboxy-terminal domain (CTD) and KOW DOMAIN-CONTAINING TRANSCRIPTION FACTOR (KTF1) which also binds RNA. INVOLVED IN DE NOVO 2 (IDN2) is supposed to assist in stabilizing the pairing between Pol V transcript and siRNA. The RDM1 protein of the DDR complex and de novo cytosine methyltransferase DOMAINS REARRANGED METHYLTRANSFERASE 2 (DRM2) binds to AGO4, bringing them close to Pol V transcribed regions and that results in DNA methylation