Fig. 3.
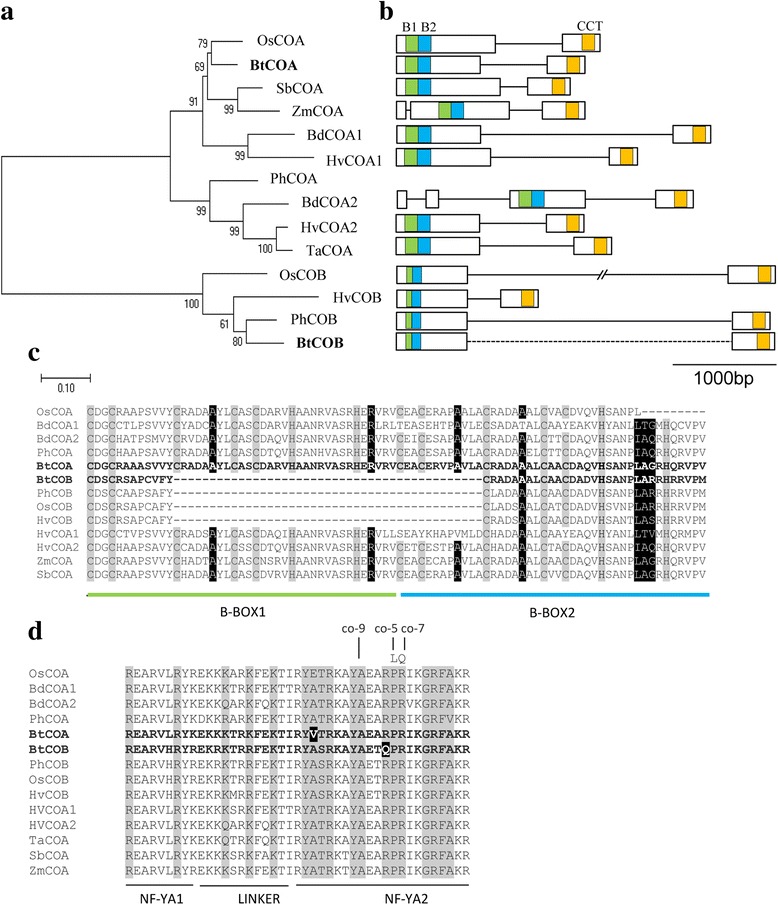
Phylogenetic and sequence characterization of BtCOA and BtCOB genes. a Phylogenetic comparison of BtCOA and BtCOB coding sequences with homologous sequences in related monocot species. The Neighbour Joining (NJ) tree was developed by Mega 7.0 using default parameters and bootstrap value 1000. b Predicted exon-intron structures of BtCOA, BtCOB genes and comparison with other monocot genes. Exons are marked as rectangles and introns as solid lines. c Multiple sequence alignment of the B-box domains of BtCOA and BtCOB protein sequences, which indicates presence of two full length B-boxes in BtCOA, while it is truncated in BtCOB. The characteristic C and H residues of B-box domains are highlighted in grey. Black highlighted amino acids are functionally important as evidenced by mutational analyses. d Sequence comparison of CCT domains between BtCOA, BtCOB and other related monocot members. Amino acids conserved for HAP3 and HAP5 binding are highlighted in grey. Amino acids not conserved in B. tulda are highlighted in black. NF-YA1 interacts with HAP3 and NF-YA2 interacts with CCAAT DNA sequences. Sequences used are: OsCOA: Os06g16370.1, BdCOA1: Bradi1g43670.1, BdCOA2: Bradi3g56260.1, PhCOA: PH01005551G0030, BtCOA: KY249523, HvCOA1: AF490467.1, HvCOA2: AF490469.1, ZmCOA: GRMZM2G405368, SbCOA: Sobic.010G115800.1, OsCOB: Os09g06464.1, PhCOB: PH01000048G0270, BtCOB: MF983714, HvCOB: AF490473.1
