Fig. 7.
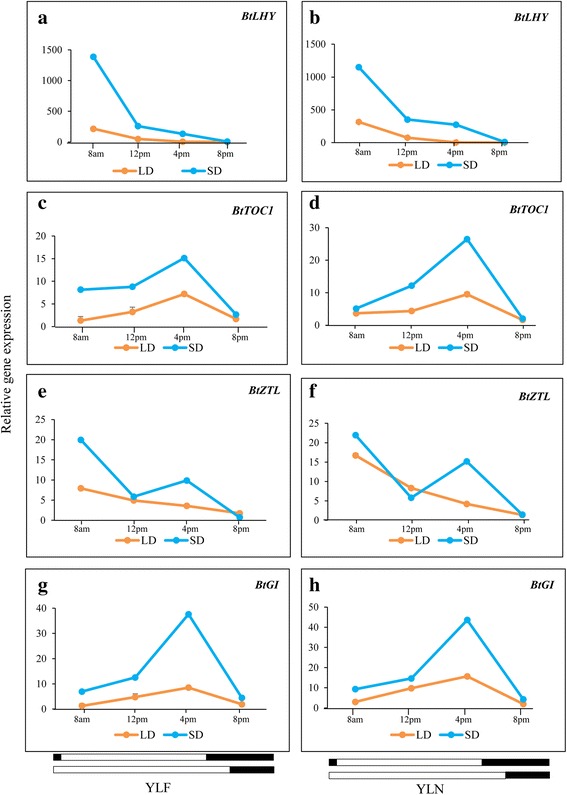
Comparison of diurnal expressions of circadian clock genes in YLF and YLN during SD and LD. a, b BtLHY, c, d BtTOC1, e, f BtZTL and g, h BtGI. Transcript expression of eIF4α was used to normalize expression data of targeted flowering genes in different tissues. The relative fold change was calculated by 2-∆∆CT method using the expression data in rhizome as calibrator and is plotted using two Y axis. Each data point in the line graph represents mean of three biological replicates ± SE in case of LD and one biological replicate in case of SD
