Fig. 5.
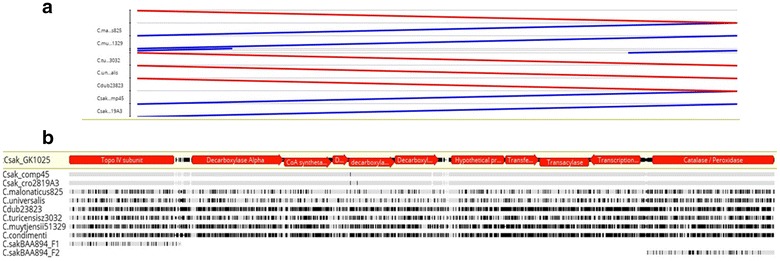
a LASTZ pairwise alignment analysis of the Cronobacter malonate utilization operon. 11.5 kb of the operon from all the listed strains were subjected to LASTZ algorithm implemented in Geneious suite. The lines indicate conserved blocks in the pairwise alignment of each operon sequence with C. sakazakii strain GK1025 sequences. The length of the lines indicates the extent of conservation ignoring coding bias. b Multiple alignment analysis of the Cronobacter malonate utilization operon. 11.5 kb of the operon from all the listed strains were aligned with the reference C. sakazakii strain GK1025 sequences. The gray horizontal bar indicate sequence similarity and black vertical bar point to possible alleles. Significant allelic differences result in clusters of black patches in some tracks. Flanking alleles from C. sakazakii strain BAA-894 F1 (gyrB) and F2 (katG) are contiguous in the genome but are separated by a 325 bp intergenic spacer. In other strains used in this illustration, the operon is seen to have inserted in this spacer region
