Fig. 5.
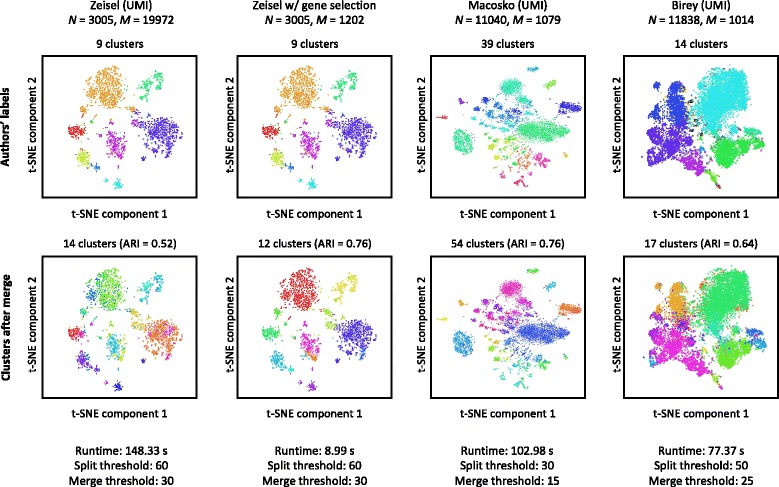
Larger datasets. DendroSplit is evaluated on three large single-cell datasets where the labels were assigned using computational methods [12, 17, 20]. Because M independent Welch’s t-tests are performed at each potential split, the runtime of the algorithm scales linearly with M. As demonstrated with the Zeisel et al. dataset, decreasing M by a factor of 16.6 likewise decreases the runtime by the same factor. The datasets here are preprocessed by filtering out genes using the procedure described by Macosko et al. [20]. This preprocessing step also improves the quality of the distance metric used, resulting in better performance
