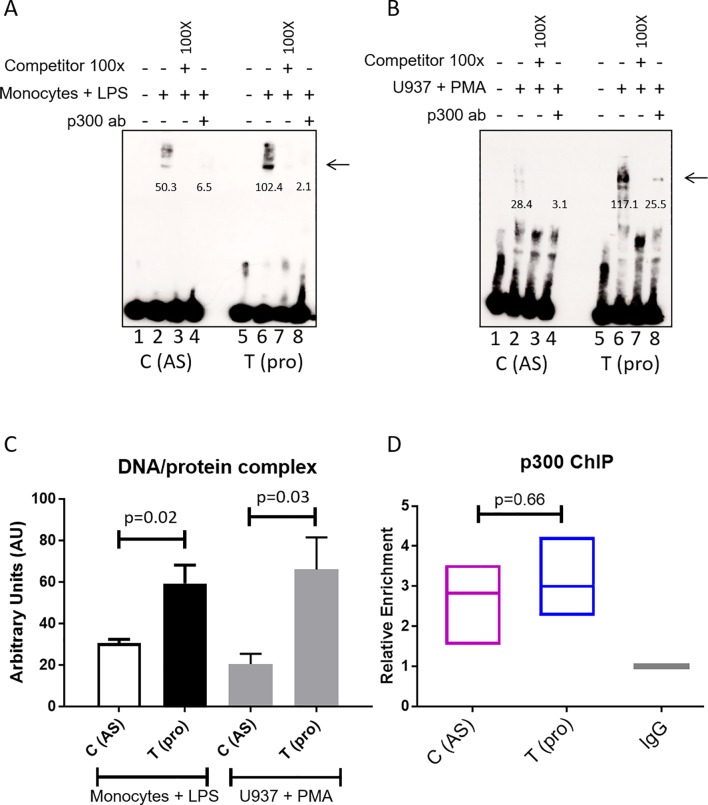
Figure 4.
rs4265380 alters protein/DNA complex formation with the involvement of p300. (A and B) Representative EMSA showing differential nuclear extract binding (complex as indicated by arrow) after addition of stimulated primary monocytes (A) and U937 cells (B) (lanes 2 and 6). The 100-fold excess of unlabelled probes has been used as competitor (lanes 3 and 7). p300 involvement was assessed by adding p300 antibody (lanes 4 and 8). Arrows indicate the presence of DNA/nuclear extract complex. Numbers below the band represent the pixel intensity measured with Image J. (C) Quantification of the DNA/protein complex of six independent EMSAs performed on both stimulated monocytes and U937 cells (P=0.019 and P=0.031, respectively, Student’s t-test). (D) Relative enrichment of p300 was assessed with chromatin immunoprecipitation experiments (n=3) on rs4265380 CT heterozygote CD14+ monocytes (after 24 hours stimulation with LPS). AS, ankylosing spondylitis; EMSA, electrophoretic mobility shift assay; LPS, lipopolysaccharide; PMA, phorbol-12-myristate-13-acetate.