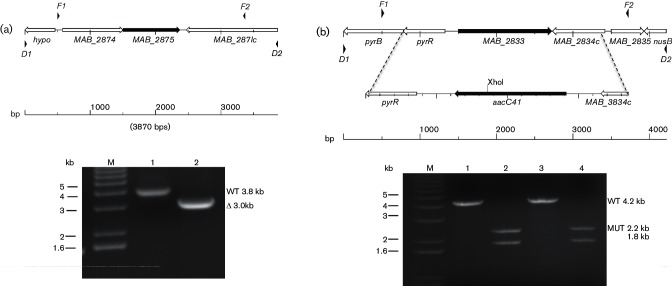
Fig. 4.
Analysis of MAB_2875 and MAB_2833 allelic exchange mutants. (a), top: map of genomic region for MAB_2875. F1 and F2 are locations for primers used to amplify region for the construction of the deletion alleles, while D1 and D2 are the locations for primers used for diagnostic PCR. (a), bottom: PCR reactions using diagnostic primers D1 and D2 for MAB_2785 and DNA prepared from: Lane 1 (PM3044 MAB_2875+ parental strain), Lane 2 (PM3386 ΔMAB_2875 mutant). (b), top: map of genomic regions for MAB_2833 with inset showing the replacement of MAB_2833 with the aacC41 apramycin resistance cassette. F1 and F2 are locations for primers used to amplify region for the construction of the deletion alleles, while D1 and D2 are the locations for primers used for diagnostic PCR. Note the unique XhoI restriction endonuclease site in the aacC41 gene inserted into the MAB_2833 deletion. Products from the MAB_2833 PCR reactions were purified and the DNA subjected to XhoI digestion prior to electrophoresis. The mutant ΔMAB_2833::aacC41 amplicon (4.0 kb) will yield two fragments of 2.2 and 1.8 kb after XhoI digestion, while the wild-type amplicon will remain unaffected at 4.2 kb. (b), bottom: ethidum bromide-stained gel of PCR reactions using diagnostic primers D1 and D2 for MAB_2833 and DNA prepared from: Lane 1 (PM3044 MAB_2833+ parental strain), Lane 2 (PM3389 ΔMAB_2833 :: aacC41 mutant), Lane 3 (PM3386 ΔMAB_2875 MAB_2833+ parental) and Lane 4 (PM3395 ΔMAB_2875ΔMAB_2833 :: aacC41 double mutant 2.2 and 1.8 kb). M is the 1 kb DNA ladder marker lane.