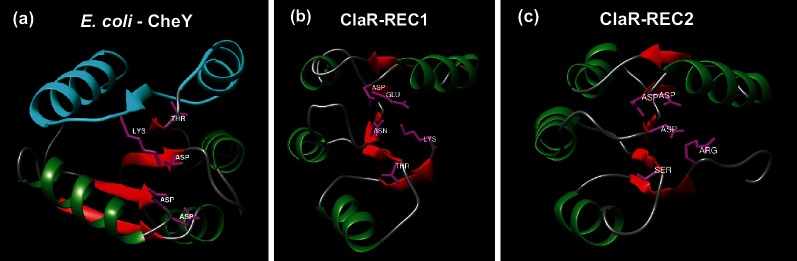
Fig. 3.
Comparison of ClaR predicted secondary structures to CheY. (a) Experimentally determined crystal structure of E. coli CheY ([41], PDB ID: 3CHY). α-Helices and β-sheets coloured in green and red, respectively. Blue colouring represents predicted CheY protein–protein interaction surface. Active site residues known to be required for phosphorylation highlighted in magenta. Structure visualized using UCSF Chimera (https://www.cgl.ucsf.edu/chimera/). (b) Predicted structure of ClaR REC1 domain (residues 1–121). Structure predicted using PHYRE2 structure prediction algorithm (http://www.sbg.bio.ic.ac.uk/phyre2) [48]. Structure colouring described in Fig. 3(a). (c) Predicted structure of ClaR REC2 domain (residues 144–252). Structure predicted also using PHYRE2 structure prediction algorithm. Structure colouring described in Fig. 3(a).