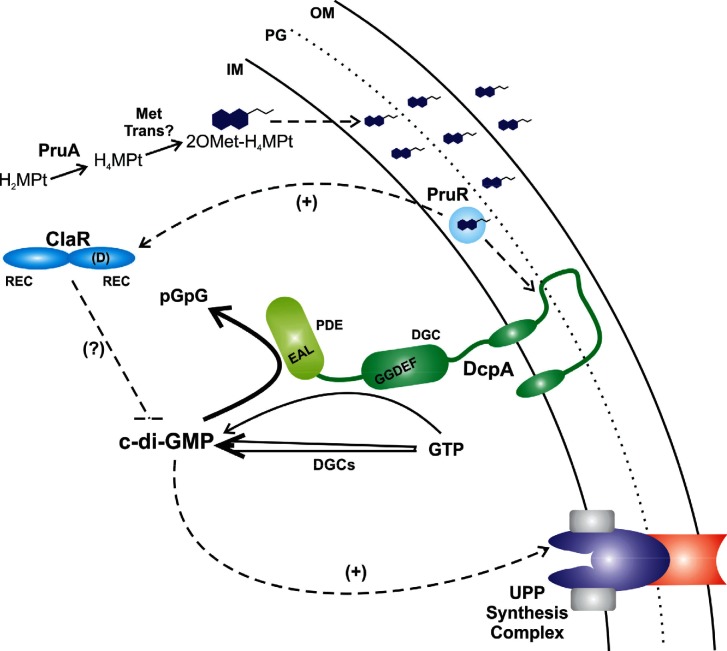
Fig. 6.
Model of ClaR regulation. The tentative model of ClaR regulation is based on data described in the figures and text, and on previously published work [30]. ClaR requires PruA and PruR, but not DcpA, to exert negative regulation of A. tumefaciens biofilm formation. The biosynthesis of the dominant PruA-dependent monapterin, 2′OMet-H4MPt, is depicted, with the compound represented as a double-ringed structure with a short acyl tail. The physical association of the monapterin species with PruR in the periplasm has not been proven, but is supported by recent work. Solid arrows are enzymatic reactions and dashed arrows are putative regulatory interactions. The ClaR dual REC domain structure with the conserved D residue is shown. Synthesis of c-di-GMP is indicated for GTP by DcpA (thin arrow) and by the other active DGCs in A. tumefaciens (double-stalked arrow) that all contribute to cellular pools of the signal. The bold arrow for c-di-GMP breakdown by DcpA is meant to show that this activity dominates under standard conditions. The Wzy-type complex is diagrammatically shown for the presumptive UPP biosynthetic machinery.