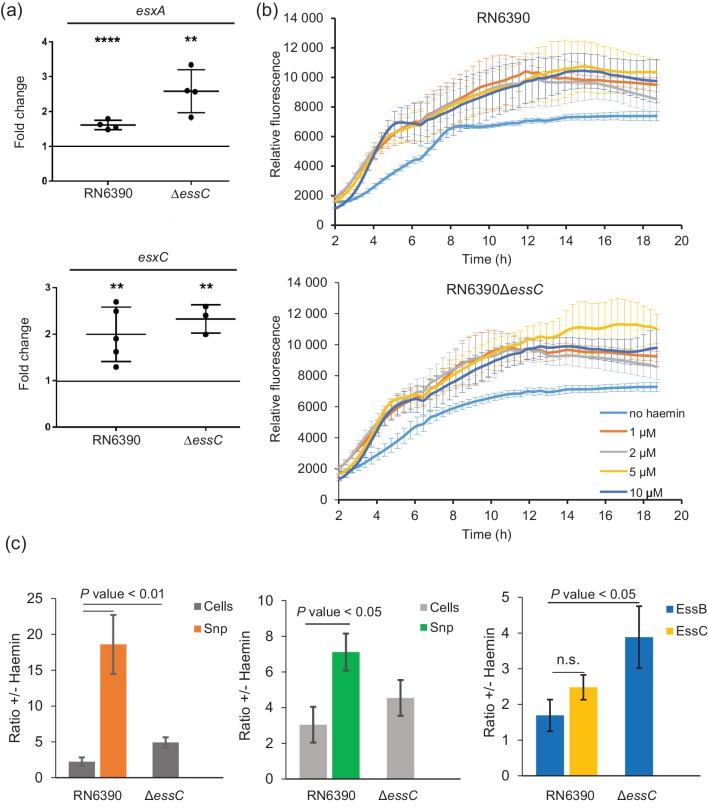
Fig. 4.
Haemin affects transcription of esxA and esxC and secretion of the encoded proteins. (a) S. aureus RN6390 and the isogenic essC deletion strain were grown aerobically in the presence or absence of 1 µM haemin to an OD600 of 1, at which point mRNA from at least three biological replicates was prepared as described in the Methods. Relative transcription levels of the esxA and esxC genes were assayed by RT-qPCR (normalized against the level of 16S rRNA). ****P<0.0001; **P<0.01. (b) S. aureus RN6390 and the isogenic essC deletion strain were cultured in the presence of the indicated concentrations of haemin for 18 h in 96-well plates (200 µl volume) with shaking. Yellow fluorescent protein (YFP) fluorescence of the culture was monitored at 485 nm and was measured in arbitrary units that were normalized to the growth at each time point. Fluorescence was statistically significantly higher in the presence of haemin than in its absence, for both RN6390 and the essC deletion strain (assessed by comparing the relative fluorescence in the absence of haemin with the relative fluorescence measured in the presence of 1, 2, 5 or 10 µM haemin at each time point sampled between 3 and 6 h) (P<0.05 in all cases). (c) Quantification of the levels of T7SS-related proteins in the presence and absence of 1 µM haemin. Cultures of RN6390 and the essC deletion strain grown aerobically in TSB medium in the presence or absence of 1 µM haemin to an OD600 of 2 and separated into cells and supernatant (sn) as described in the Methods. For quantification of protein in the cellular fraction, 5 µl of a cell culture adjusted to an OD600 of 1 was loaded and for the TCA-precipitated supernatant an equivalent of 15 µl of culture supernatant was loaded. Quantification results are expressed as the ratio of the signal obtained in the presence 1 µM haemin to that in the absence of haemin. The results represent the mean±sd of at least three biological replicates. P values from Student's paired t-tests are shown.