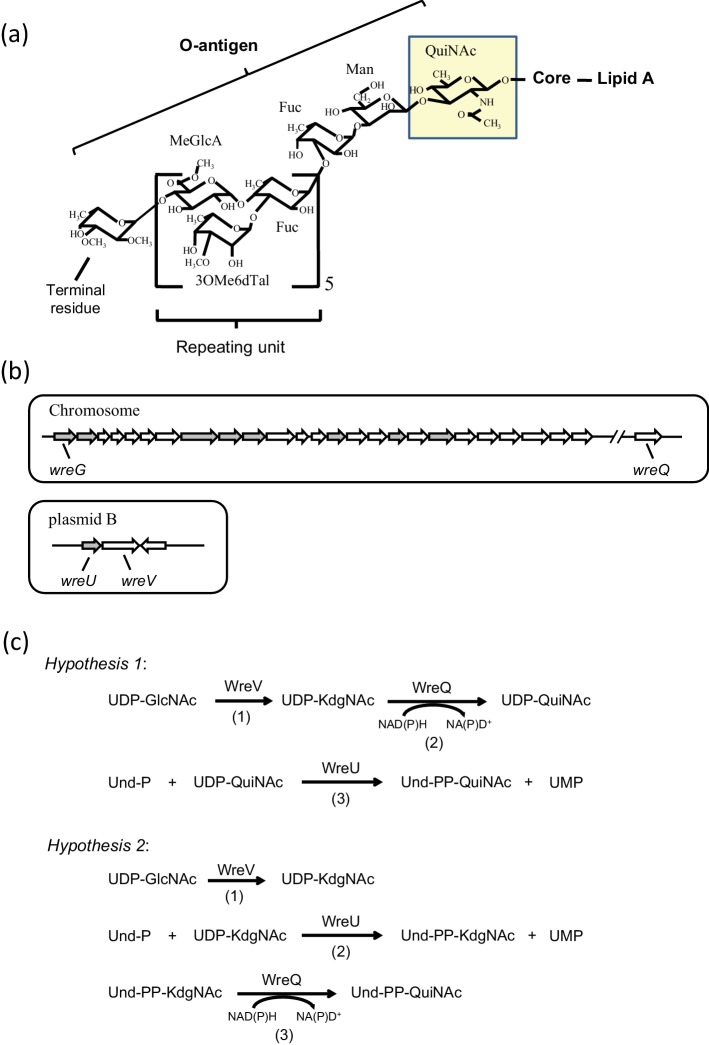
Fig. 1.
(a) Structure of Rhizobium etli CE3 O-antigen. The O-antigen structure of R. etli LPS is shown linked to the lipid A core. Abbreviations for the sugars: QuiNAc, N-acetyl-d-quinovosamine; Man, mannose; Fuc, fucose; MeGlcA, methyl-glucuronate; 3OMe6dTal, 3-O-methyl-6-deoxytalose; terminal residue, TOMFuc, 2,3,4-tri-O-methylfucose or DOMFuc, 2,3-di-O-methylfucose. The proposed first O-antigen sugar, QuiNAc, is highlighted. (b) R. etli CE3 O-antigen genetic clusters. Upper panel: the chromosomal wre gene cluster (previously called lps region α) spanning nucleotides 784 527 to 812 262 of the genome sequence consists of 25 predicted ORFs. Another chromosomal ORF (wreQ) spanning nucleotides 2 969 313 to 2 970 242 is required for QuiNAc synthesis [27]. Lower panel: a 4-kilobase cluster on plasmid pCFN42b consists of three predicted ORFs. In each panel the predicted GTase-encoding genes are in grey. Genes encoding enzymes studied in the current work, wreG, wreQ, wreU and wreV, are specifically labelled. (c) Two hypotheses of O-antigen initiation in R. etli CE3. Reactions in each hypothesis and the enzyme that catalyses each reaction are indicated. In both hypotheses, the first reaction (1) is the same, the conversion of UDP-GlcNAc to UDP-KdgNAc catalysed by the predicted 4,6-dehydratase WreV. The two hypotheses differ in reactions (2) and (3). In hypothesis 1, KdgNAc is reduced to QuiNAc on the UDP linkage by WreQ and then QuiNAc-1-P is transferred by WreU. In hypothesis 2, KdgNAc-1-P is transferred by WreU first and then KdgNAc is reduced to QuiNAc by WreQ on the Und-PP linkage.