Figure 3.
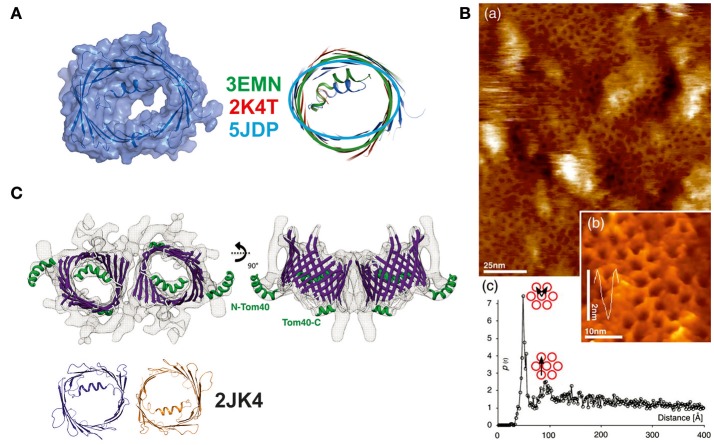
New structural details collected on members of the VDAC-superfamily of 19-stranded β-barrels. (A) Structure determination of the E73V mutant of human VDAC. This mutant was previously described to have altered biophysical properties in comparison to the wildtype protein. The NMR determination of the structure yielded a structurally significantly changed beta-barrel with an oval rather than a circular form and a smaller channel diameter. Three structures of the mVDAC1 (in green), hVDAC1 (in red), and the mutant structure (in light blue) were superimposed to show the deviation in the barrel section. A surface representation of the latest NMR structure (5JDP) shows the particular small diameter of the pore eyelet in E73V (Jaremko et al., 2016). (B) (a) High resolution AFM topographs of densely packed scVDAC natively embedded in membranes of S. cerevisiae (large picture) and (b) high resolution view of VDAC molecules in an arrangement similar to the early electron microsocopy images (inset picture). (c) Peak analysis of the separation between two VDAC molecules yields a distance of 53 Å (pore to pore; see graph) (Gonçalves et al., 2007). (C) Structural analysis of the Tom40 complex shows a dimeric arrangement of the pore structures with the alpha-helix inside the barrel or exposed to the surface. The structure at 1 nm resolution was modeled by a Tom40-homology model based on the mVDAC structure (PDB-entry 3EMN) and can accommodate the 19 beta-strands unambiguously. The dimeric arrangement is mediated by the same beta1/beta19 strands as previously reported for the hVDAC1 dimer (2JK4—dimer shown in orange and dark blue) (Bayrhuber et al., 2008; Bausewein et al., 2017). Figures reproduced with permission.