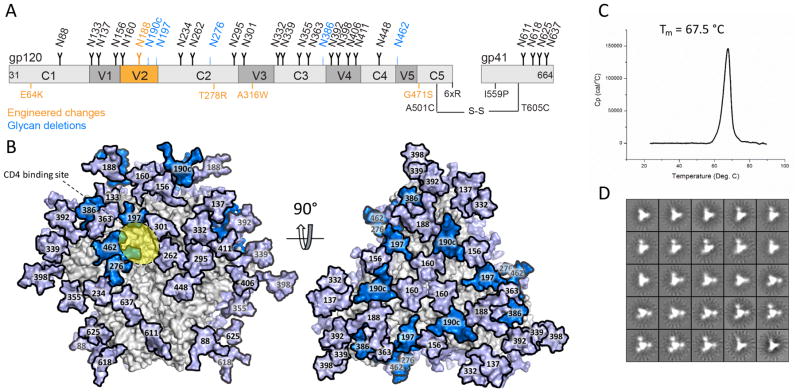
Figure 1. Design and properties of CHO-cell produced BG505 SOSIP.v4.1-GT1 trimers.
(A) Schematic representation of the BG505 SOSIP.v4.1-GT1 construct. The constant (C1–C5) and variable (V1–V5) regions of gp120 are highlighted in light and dark grey, respectively. The stabilizing substitutions present in the SOSIP.664 are shown in black, with modifications that create the SOSIP.v4.1-GT1 construct in orange. Of note is that the E64K and A316W mutations were introduced to create the BG505 SOSIP.v4.1 trimer (49). Detailed information on the GT1 construct, including a 7 amino acid deletion in V2, are described elsewhere (63). The SOSIP.664 glycans are shown as black icons, with the ones deleted from the GT1 construct colored blue. Glycan N188 (orange) in the altered V2 region is equivalent to glycan N190 on the prototype BG505 SOSIP.664 version, and thus represents neither a glycan introduction nor a deletion. (B) The glycans deleted from the GT1 trimer are highlighted in marine blue on a model of the fully glycosylated BG505 SOSIP.664 trimer (the remaining glycans are in light blue). The approximate location of the CD4bs is highlighted in yellow. The model was constructed using PDB ID 5ACO (80) as previously described (41). (C) DSC analysis of 2G12 plus SEC-purified GT1 trimers, with the Tm value indicated (see Table S1 for additional relevant Tm values). (D) NS-EM analysis of the same purified GT1 trimers, which were 100% native-like.