Figure 1.
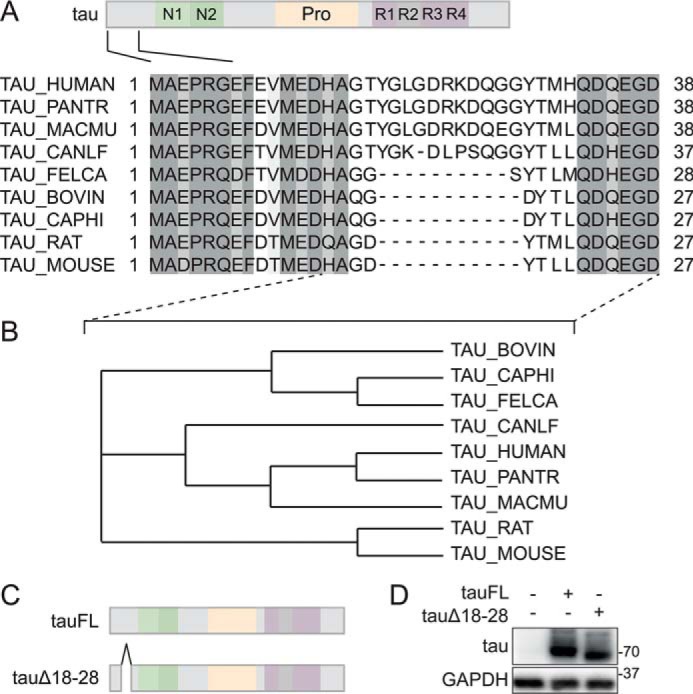
A unique 11-amino acid motif in the N-terminal region of primate tau. A, alignment of mammalian tau primary sequences. Primary sequences of N-terminal regions of human, non-human primate, macaque, bovine, goat, dog, cat, and rodent tau proteins were aligned using ClustalOmega with marked up similarities (gray shading). Note the absence of a motif between amino acids 18 and 28 of primate tau in other mammalian tau sequences. Only canine tau contains a 10-amino acid motif of lesser similarity to primate tau. B, dendrogram of N-terminal mammalian tau protein sequences. Dendrogram analysis of sequences between the indicated residues in A (dashed lines) was performed using ClustalOmega. Note that canine tau clusters with primate (human, Pan troglodytes, and Macaca mulatta) tau N-terminal sequences due to its 10-amino acid motif albeit with weaker similarity. C, amino acids 18–28 were deleted in human tau40 to generate the tauΔ18 construct. D, tauΔ18 and tau full-length (tauFL) were transfected into 293T cells. Immunoblot of cell lysates from tauFL, tauΔ18, or untransfected control was detected with anti-human tau antibody. tauΔ18 shows reduced retention, consistent with deletion of amino acids 18–28. Probing for glyceraldehyde-phosphate dehydrogenase (GAPDH) served as loading control.
