Abstract
Background:
Acinetobacter baumannii, is an emerging nosocomial multidrug resistance pathogen with the rapid spread of clones being reported in health-care settings and hospitals worldwide. Carbapenem resistance in this bacterium has been attributed to D OXA β-lactamases with OXA-51-like β-lactamase, being present in all A. baumannii isolate. The present study looks into the antibiotics susceptibility and molecular characterization of clinical A. baumannii isolates from Intensive Care Unit (ICU) samples in Al-Hofuf, South-eastern region of Saudi Arabia.
Materials and Methods:
Eleven strains of ICU A. baumanni i isolates were used for the investigation. Bacteria isolation was by basic microbiological techniques. Organisms identification and antibiogram susceptibility testing was by the BioMerieux VITEK 2 compact automated system (BioMerieux, Marcy I'Etoile France), according to the manufacturers guidelines. Confirmation of A. baumannii was by the presence of the OX-51 gene, also, carbapenemase encoding resistant genesblaOXA-23, blaOXA-40, and blaOXA-51, were analyzed using multiplex PCR. The Student's t test was used to analyze the obtained data for between group comparisons with statistically significance level set at P < 0.05.
Results:
Eight of the isolates were confirmed to be A. baumannii. Five of which were resistant to the carbapenems against which they had been tested. One isolate was resistant to tigecycline, whereas three tested intermediate to the drug. OXA-23 was detected in isolates 1, 4, 5, 6, and 7.
Conclusion:
It can, therefore, be concluded that the probable predominate carbapenems resistant genes in ICU isolates from the present investigation, are those associated with OXA-23.
Keywords: Acinetobacter baumannii, Intensive Care Unit, multidrug resistance, OXA-23, OXA-51, extensive drug resistance
Introduction
The increase in difficult to treat multi-drug resistant bacteria has created an unprecedented problem in the 21st century. Acinetobacter baumannii has been listed among the most difficult antimicrobial resistant Gram-negative bacilli.[1,2] A. baumannii an opportunistic pathogen has evolved to be an important pathogen associated with nosocomial infections worldwide and is known to have developed strains that are multidrug resistance (MDR), extensive drug resistance (XDR) and Pandrug resistance.[3] Researchers [4,5,6,7,8] have carried out extensive investigations into various aspects of this bacteria supper bug in which they had looked into the various genes responsible for carbapenemase resistance. A. baumannii is postulated to have emerged as a nosocomial pathogen with the rapid spread of clones being reported in hospitals and health-care settings [8,9,10] Outbreaks of MDR A. baumannii in critical care units have also been reported in various regions of the world.[5,11] Furthermore, colonization with this bacterium in immunocompromised patients have been reported by researchers [6,12] with diabetes being shown to be a significant risk factor for A. baumannii nosocomial infections.[6,13,14] Other risk factors which have been associated with patient colonization by A. baumannii are exposure to ICUs as well as prolonged hospital stays.[1] Patients at risk could be through exposure to mechanical ventilation, recent surgery, invasive procedures [15,16] as well as items used in the care of patients.[11,17]
The carbapenems had been the drug of choice in treatment due to their effective antibacterial activity as well as having low toxicity. However, A. baumannii has emerged resistant to this drug of choice resulting in a major global health concern.[18,19] Resistance to the carbapenem manifested in plasmid-encoded β-lactamase (OXA-23, OXA-40, and OXA-58) enzymes and the chromosomally encoded OXA-51.[20] The clones harbored by different A. baumannii isolates have been shown to differ.[20] Differences have been indicated between the isolates diabetic and nondiabetic patients [6] with no specifics indicating if there are clones associated with diabetes. The present work looks into A. baumannii isolates from Intensive Care Unit (ICU) samples with a view of providing an insight in the prevalent OXA β-lactamase associated with ICU isolates. This would help provide information on the genes associated with ICU isolates as there is the need for constant surveillance in the microbial susceptibility of bacteria isolates in this era of evolving bacteria superbugs.
Materials and Methods
Sample collection and processing
Clinical isolates on A. baumannii obtained from ICU samples were used for the study. Samples had been isolated from sputum, throat, abdomen, catheter and endotracheal aspirates and plated out on MacConkey agar. A. baumannii isolation and identification was carried out in hospitals using basic microbiological and biochemical techniques.
Isolation was through basic microbial techniques while confirmation of isolates was carried out using BioMerieux VITEK 2 compact automated system (BioMerieux, Marcy I'Etoile France), according to the manufacturer's guidelines. For further confirmation, PCR was used to detect the intrinsic blaOXA-51- gene.[6]
Antimicrobial susceptibility testing
Pure samples grown overnight on MacConkey agar were used for the antibiogram testing. The antibiotic susceptibility testing was carried out using the VITEK 2 compact automated system according to manufacturer's guidelines. Isolates were tested against the GN cards with the following antibiotics: cefazolin, imipenem, meropenem, gentamicin, tobramycin, ciprofloxacin, levofloxacin, tigecycline, and sulfamethoxazole/trimethoprim. Only isolates which showed resistance to imipenem and meropenem were used for the investigations. The minimum inhibitory concentration values were determined using the ETest strips (AB Biodisks) following the manufacturer's guidelines. The test experiments were repeated in triplicates.
Molecular analysis for carbapenem-resistant genes
The presence of carbapenemase encoding resistant gene (blaOXA-23, blaOXA-40, blaOXA-51) among the isolates were detected using multiplex PCRgenes using primers pairs.[8,21,22,23] The primers used for the amplification and sequencing are shown in Table 1.
Table 1.
Sequence of primers used for the study

The PCR protocol is as described by Marco et al.[21] Samples were initially denatured for 5 min at 94°C, followed by amplification cycles and elongation steps as described by Marco et al.[21] Resulting PCR products were visualized after agarose gel electrophoresis, staining with ethidium bromide.
Statistical analysis
Excel software, 2013 version was used to analyze the obtained data, with student's T-test used for comparison between groups. Statistically, significance level was set at P < 0.05.
Results
A total of eight nonrepetitive confirmed isolates of A. baumannii collected from ICUs in the study region were used for the investigation. The isolate source and the minimum inhibitory concentration (MIC) values are shown in Table 2. The result on the susceptibility of these isolates against tested antibiotics is presented in Table 3. The isolates are seen to exhibit varying degrees of resistance to the different antibiotics with 56% of them being resistant to imipenem and meropenem. However, all (100%) the isolates were sensitive to colistin, One (12.5%) isolate was resistant to tigecycline, whereas 33.33% of the isolates were intermediate to tigecycline. Based on their resistance against the tested antibiotic groupings, 67% of the isolates are MDR. Figure 1 shows the results of the resistance of these isolates to the antibiotic categories. Highest resistance was against the cephalosporins (78%) followed by fluoroquinolones with a 67% resistance by the isolates. Fifty-six percent of the A. baumannii were each resistant to the carbapenems and penicillin, whereas 44% of them were each resistant to the aminoglycosides and trimethoprim/sulfamethoxazole.
Table 2.
Source of isolation and the minimum inhibitory concentration vales against tested antibiotics
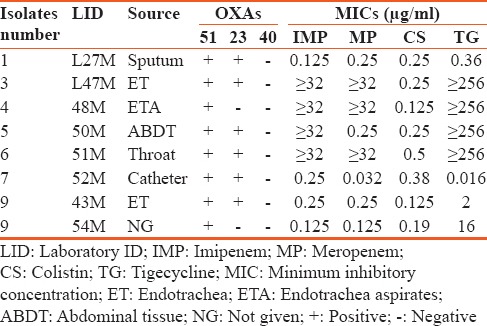
Table 3.
Antibiogram of multidrug resistance clinical Acinetobacter baumannii isolates from Intensive Care Units
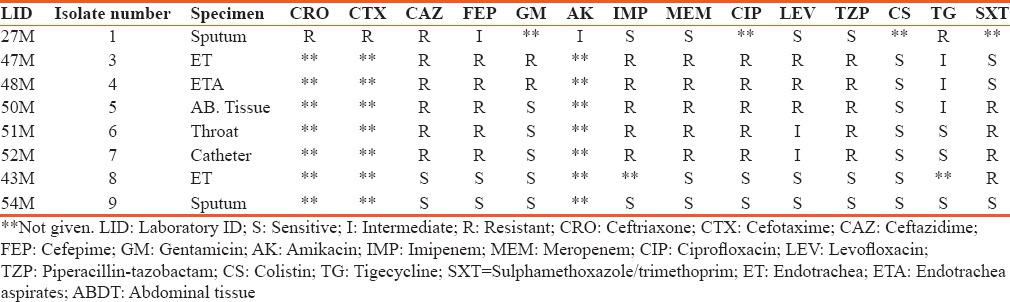
Figure 1.
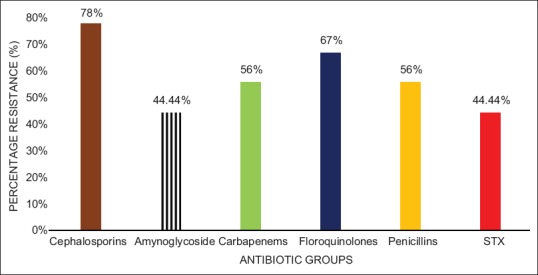
Percentage resistance of Acinetobacter baumannii to anti-microbial categories. STX = Sulphamethoxazole/trimethoprim
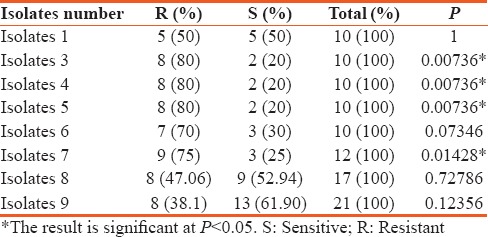
A comparison between the number of antibiotics that the isolates were resistant and sensitive to is shown in Table 4. For isolate 1, there was about a 50% resistance against the tested antibiotics. The difference between the number of antibiotics that the isolate was resistant or sensitive to is not statistically significant. However, for isolates 3, 4, 5, and 7, there was a 75% to 80% resistance against the tested antimicrobials, inclusive of the carbapenems. The results are statistically significant (P < 0.05). Isolates 8 and 9 were more susceptible to most of the tested drugs and the difference against the drugs to which they were resistant to was not statistically significant as shown in Table 4.
Table 4.
Comparison between resistance and sensitivity against the tested antibiotics by Intensive Care Unit isolates of Acinetobacter baumannii
On the other hand, for the comparison between the antibiotics to which isolates were resistant or intermediate to, results showed that there is a statistically significant difference (P < 0.05) with the exception of isolate 1 that recorded no statistical difference [Table 5]. Also, the results on the prevalent genes seen to be associated with the ICU A. baumannii isolates is shown in Figure 2. The OXA-23 was present in 56% of the isolates while OXA 40 was not encountered in any of the isolates.
Table 5.
Relationship between intermediate and resistance to tested antibiotics among Intensive Care Unit Acinetobacter baumannii isolates
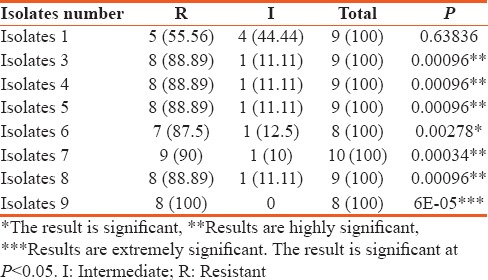
Figure 2.
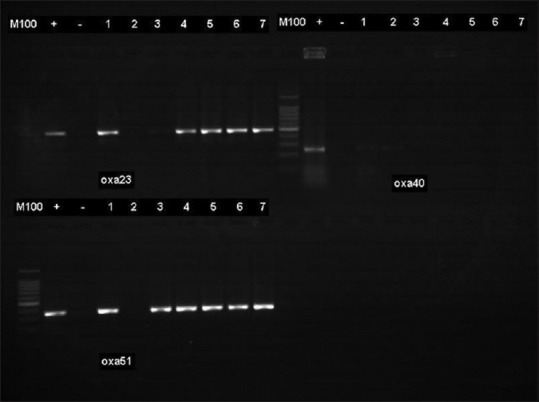
Characterisation of carbapenemase genes. The OXA-23 enzyme was detected in 5 (56%) of the isolates. All the isolates were negative to OXA-40
Discussion
The health-care problem caused by MDR A. baumannii is further highlighted in the results of the present investigation. The isolation of MDR A. baumannii from ICU samples as seen in the present study is not unexpected as such isolations had been reported earlier by researchers.[7,15,16,22] Furthermore, the evolvement of both MDR and XDR A. baumannii has been reported by Kempf and Rolain in 2012.[23]
The results from the present study also showed majority of the isolates to be MDR, whereas >50% of them showed XDR against the tested antibiotic groups. The exhibition of high levels of MDR and XDR by A. baumannii isolates has been reported in different regions of the world. Nageeb et al.[22] reported a 100% MDR and 80% XDR among A. baumannii isolates obtained from ICU in Egypt, whereas in Pakistan, Evans et al.,[24] encountered 82.4% MDR and 65% XDR among the isolates in their study. Both findings differ from those of the present investigation. However, Dent et al.,[25] reported a 72% and 58% MDR and XDR, respectively, among A. baumannii in their study Nashville General Hospital and this finding is similar to those of the present study. While being certain that all the A. baumannii ICU isolates in the present study showed both MDR and XDR against the tested antibiotic groups, the observed difference between researches could be attributed to a number of contributing factors one of which is geographical regional isolate differences, as well as the total number of isolates being used for the investigations by the different researchers as had been indicated by Nageeb et al.[22]
Furthermore, the resistance by more than half of the isolates to the carbapenems and other β-lactams as seen in the present investigation indicates that carbapenem resistance remains a problem. Al-Sultan et al.[6] had reported that carbapenem resistance was very high among A. baumannii hospitals isolates in Saudi Arabia. The OXA-51- and OXA-23-like genes were the encountered genes responsible for carbapenem resistance in the A. baumannii isolates in the present study as shown in Figure 2. Yaowen et al.[26] observed OXA-23 gene to be the major carbapenemase responsible for resistance in their isolates from a teaching hospital. The findings are also consistent with those of Merino et al.[27] and Zowawi et al.[28] Observed in these carbapenem-resistant ICU isolates was the carriage of both the OXA-23 and OXA-51 genes. Similar findings had been reported by Al-Sultan et al.,[6] in 2013 on A. baumannii isolates from diabetic patients. The presence of both genes was however detected in one isolate which was susceptible to both imipenem and meropenem. This isolate was also susceptible to other β-lactams, fluoroquinolone and intermediate to aminoglycoside. Similar to this finding is that of Yaowen et al.,[26] that reported an A. baumannii isolate which had been susceptible to both imipenem and meropenem but was found to be carrying the blaOAX-72 gene with no other resistant gene. This isolate was reported as susceptible to other β-lactams, aminoglycosides, and fluoroquinolones among other antibiotics.[29] The authors concluded however that there was a need for more study to find out why the presence of OXA-72 did not produce β-lactam resistance. Furthermore, El-Shazy et al.,[29] in 2015 reported that not all the A. baumannii isolates in their study harboring the ISAbla1 gene were resistant to the carbapenems. Evans et al.[24] found the OXA-51-like enzyme expressed in a sensitive A. baumannii isolates and explained that the enzyme demonstrated the capability to dramatically increase the MIC to carbapenem for an isolate. They also indicated that the OXA-51-like enzyme demonstrated only a weak hydrolytic activity against carbapenemase. These might therefore explain the reason for the results recorded with one of the isolate in the present study. However, there is need for further study on the nonconferring of resistance in the presence of OXA-like gene as earlier suggested.[27]
Conclusion
From the results of the present investigation, it can be concluded that the predominant carbapenems resistant in the ICU A. baumannii isolates in the present investigation, are those of the OXA-23. There is however need for more investigation for both monitoring and further updating.
Financial support and sponsorship
Nil.
Conflicts of interest
There are no conflicts of interest.
Acknowledgment
The authors would like to thank Dr. Abdulrahman Al-Sultan for his expertise advice, Mr. Hani Al-Rasasi and Ms. Hajer, for their technical assistance.
References
- 1.Maragakis LL, Perl TM. Acinetobacter baumannii: Epidemiology, antimicrobial resistance, and treatment options. Clin Infect Dis. 2008;46:1254–63. doi: 10.1086/529198. [DOI] [PubMed] [Google Scholar]
- 2.Boucher HW, Talbot GH, Bradley JS, Edwards JE, Gilbert D, Rice LB, et al. Bad bugs, no drugs: No ESKAPE! An update from the Infectious Diseases Society of America. Clin Infect Dis. 2009;48:1–2. doi: 10.1086/595011. [DOI] [PubMed] [Google Scholar]
- 3.Christopher AF, Hora S, Ali Z. Investigation of plasmid profile, antibiotic susceptibility pattern multiple antibiotic resistance index calculation of Escherichia coli isolates obtained from different human clinical specimens at tertiary care hospital in Bareilly - India. Ann Trop Med Public Health. 2013;6:285–9. [Google Scholar]
- 4.Senok A, Garaween G, Raji A, Khubnani H, Kim Sing G, Shibl A, et al. Genetic relatedness of clinical and environmental Acinetobacter baumanii isolates from an intensive care unit outbreak. J Infect Dev Ctries. 2015;9:665–9. doi: 10.3855/jidc.6726. [DOI] [PubMed] [Google Scholar]
- 5.Ushizawa H, Yahata Y, Endo T, Iwashima T, Misawa M, Sonobe M, et al. A epidemiological investigation of a nosocomial outbreak of multidrug-resistant Acinetobacter baumannii in a critical care center in Japan, 2011-2012. Jpn J Infect Dis. 2016;69:143–8. doi: 10.7883/yoken.JJID.2015.049. [DOI] [PubMed] [Google Scholar]
- 6.Alsultan AA, Evans BA, Elsayed EA, Al-Thawadi SI, Al-Taher AY, Amyes SG, et al. High frequency of carbapenem-resistant Acinetobacter baumannii in patients with diabetes mellitus in Saudi Arabia. J Med Microbiol. 2013;62:885–8. doi: 10.1099/jmm.0.057216-0. [DOI] [PubMed] [Google Scholar]
- 7.El-Ageery SM, Abo-Shadi MA, Alghaithy AA, Ahmad MA, Alsharif NH, Alharbi SA, et al. Epidemiological investigation of nosocomial infection with multidrug-resistant Acinetobacter baumannii. Eur Rev Med Pharmacol Sci. 2012;16:1834–9. [PubMed] [Google Scholar]
- 8.Qi C, Malczynski M, Parker M, Scheetz MH. Characterization of genetic diversity of carbapenem-resistant Acinetobacter baumannii clinical strains collected from 2004 to 2007. J Clin Microbiol. 2008;46:1106–9. doi: 10.1128/JCM.01877-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jean SS, Hsueh PR, Lee WS, Chang HT, Chou MY, Chen IS, et al. Nationwide surveillance of antimicrobial resistance among non-fermentative Gram-negative bacteria in Intensive Care Units in Taiwan: SMART programme data 2005. Int J Antimicrob Agents. 2009;33:266–71. doi: 10.1016/j.ijantimicag.2008.08.026. [DOI] [PubMed] [Google Scholar]
- 10.Chen CM, Liu PY, Ke SC, Wu HJ, Wu LT. Investigation of carbapenem-resistant Acinetobacter Baumannii isolates in a district hospital in Taiwan. Diagn Microbiol Infect Dis. 2009;63:394–7. doi: 10.1016/j.diagmicrobio.2008.12.011. [DOI] [PubMed] [Google Scholar]
- 11.Wilks M, Wilson A, Warwick S, Price E, Kennedy D, Ely A, et al. Control of an outbreak of multidrug-resistant Acinetobacter baumannii-calcoaceticus colonization and infection in an intensive care unit (ICU) without closing the ICU or placing patients in isolation. Infect Control Hosp Epidemiol. 2006;27:654–8. doi: 10.1086/507011. [DOI] [PubMed] [Google Scholar]
- 12.Navon-Venezia S, Leavitt A, Carmeli Y. High tigecycline resistance in multidrug-resistant Acinetobacter baumannii. J Antimicrob Chemother. 2007;59:772–4. doi: 10.1093/jac/dkm018. [DOI] [PubMed] [Google Scholar]
- 13.Prata-Rocha ML, Gontijo-Filho PP, Melo GB. Factors influencing survival in patients with multidrug-resistant Acinetobacter baumannii infection. Braz J Infect Dis. 2012;16:237–41. [PubMed] [Google Scholar]
- 14.Danaei G, Finucane MM, Lu Y, Singh GM, Cowan MJ, Paciorek CJ, et al. National, regional, and global trends in fasting plasma glucose and diabetes prevalence since 1980: Systematic analysis of health examination surveys and epidemiological studies with 370 country-years and 2·7 million participants. Lancet. 2011;378:31–40. doi: 10.1016/S0140-6736(11)60679-X. [DOI] [PubMed] [Google Scholar]
- 15.Fournier PE, Richet H. The epidemiology and control of Acinetobacter baumannii in health care facilities. Clin Infect Dis. 2006;42:692–9. doi: 10.1086/500202. [DOI] [PubMed] [Google Scholar]
- 16.Playford EG, Craig JC, Iredell JR. Carbapenem-resistant Acinetobacter baumannii in intensive care unit patients: Risk factors for acquisition, infection and their consequences. J Hosp Infect. 2007;65:204–11. doi: 10.1016/j.jhin.2006.11.010. [DOI] [PubMed] [Google Scholar]
- 17.Bernards AT, Harinck HI, Dijkshoorn L, van der Reijden TJ, van den Broek PJ. Persistent Acinetobacter baumannii? Look inside your medical equipment. Infect Control Hosp Epidemiol. 2004;25:1002–4. doi: 10.1086/502335. [DOI] [PubMed] [Google Scholar]
- 18.Tiwari V, Kapil A, Moganty RR. Carbapenem-hydrolyzing oxacillinase in high resistant strains of Acinetobacter baumannii isolated from India. Microb Pathog. 2012;53:81–6. doi: 10.1016/j.micpath.2012.05.004. [DOI] [PubMed] [Google Scholar]
- 19.Tiwari V, Vashistt J, Kapil A, Moganty RR. Comparative proteomics of inner membrane fraction from carbapenem-resistant Acinetobacter baumannii with a reference strain. PLoS One. 2012;7:e39451. doi: 10.1371/journal.pone.0039451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Alsultan AA, Hamouda A, Evans BA, Amyes SG. Acinetobacter baumannii: Emergence of four strains with novel bla (OXA-51-like) genes in patients with diabetes mellitus. J Chemother. 2009;21:290–5. doi: 10.1179/joc.2009.21.3.290. [DOI] [PubMed] [Google Scholar]
- 21.Dettori M, Piana A, Deriu MG, Lo Curto P, Cossu A, Musumeci R, et al. Outbreak of multidrug-resistant Acinetobacter baumannii in an intensive care unit. New Microbiol. 2014;37:185–91. [PubMed] [Google Scholar]
- 22.Nageeb W, Kamel M, Zakaria S, Metwally L. Phenotypic characterization of Acinetobacter baumannii isolates from intensive care units at a tertiary-care hospital in Egypt. East Mediterr Health J. 2014;20:203–11. [PubMed] [Google Scholar]
- 23.Kempf M, Rolain JM. Emergence of resistance to carbapenems in Acinetobacter baumannii in Europe: Clinical impact and therapeutic options. Int J Antimicrob Agents. 2012;39:105–14. doi: 10.1016/j.ijantimicag.2011.10.004. [DOI] [PubMed] [Google Scholar]
- 24.Evans BA, Hamouda A, Abbasi SA, Khan FA, Amyes SG. High prevalence of unrelated multidrug-resistant Acinetobacter baumannii isolates in Pakistani military hospitals. Int J Antimicrob Agents. 2011;37:580–1. doi: 10.1016/j.ijantimicag.2011.01.023. [DOI] [PubMed] [Google Scholar]
- 25.Dent LL, Marshall DR, Pratap S, Hulette RB. Multidrug resistant Acinetobacter baumannii: A descriptive study in a city hospital. BMC Infect Dis. 2010;10:196. doi: 10.1186/1471-2334-10-196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chang Y, Luan G, Xu Y, Wang Y, Shen M, Zhang C, et al. Characterization of carbapenem-resistant Acinetobacter baumannii isolates in a Chinese teaching hospital. Front Microbiol. 2015;6:910. doi: 10.3389/fmicb.2015.00910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Merino M, Poza M, Roca I, Barba MJ, Sousa MD, Vila J. et al.Nosocomial outbreak of a multi-resistant Acinetobacter baumannii expressing OXA-23 carbapenemase in Spain. Microb Drug Resist. 2014;20:259–63. doi: 10.1089/mdr.2013.0127. [DOI] [PubMed] [Google Scholar]
- 28.Zowawi HM, Sartor AL, Sidjabat HE, Balkhy HH, Walsh TR, Al Johani SM, et al. Molecular epidemiology of carbapenem-resistant Acinetobacter baumannii isolates in the Gulf Cooperation Council States: Dominance of OXA-23-type producers. J Clin Microbiol. 2015;53:896–903. doi: 10.1128/JCM.02784-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.El-Shazly S, Dashti A, Vali L, Bolaris M, Ibrahim AS. Molecular epidemiology and characterization of multiple drug-resistant (MDR) clinical isolates of Acinetobacter baumannii. Int J Infect Dis. 2015;41:42–9. doi: 10.1016/j.ijid.2015.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]


