Figure 7.
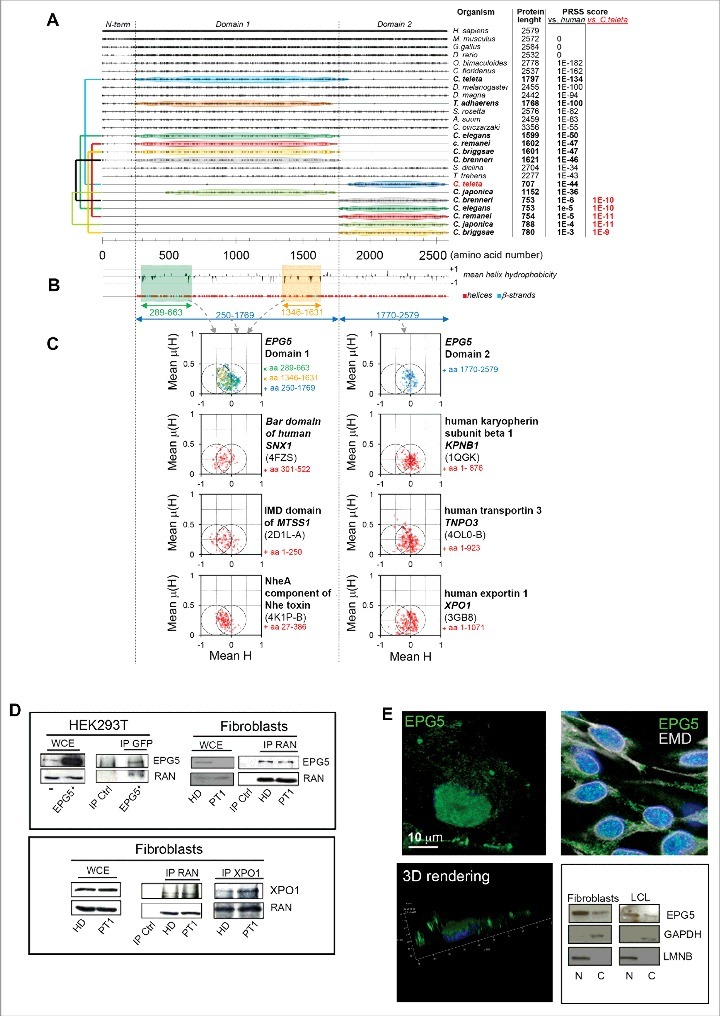
Conservation analysis, predicted secondary structures, and hydrophobic moment plots of the EPG5 protein. (A) Plot of the amino acid identities between EPG5 protein from various organisms and human EPG5 protein as obtained from multiple sequence alignment. For each sequence, PRSS scores are reported (versus the human protein), and for proteins showing poor homology (PRSS scores ≤ 10-6) these scores were also calculated versus the Capitella teleta protein and indicated in red. The colored connectors join pairs of non-overlapping sequences (named domain 1 and domain 2) found for the same organism and encompassing, taken together, the full structured region of EPG5 as inferred from secondary structure predictions by PSIPRED (shown in [B] together with the plot of the mean hydrophobicity calculated with a sliding 11-residue-long window). (C) The hydrophobic moment plots calculated for different residue ranges of human EPG5, and, for comparison, also for the known helical regions of protein domains involved in membrane remodeling (the Bar domain of human SNX1, the F-actin binding domain IMD of mim, and the NheA component of Nhe toxin) and of some characterized karyopherins (human KPNB1, human TNPO3, and human XPO1). The Protein Data Bank accession codes used to extract the information of the helical regions for these proteins are indicated in parentheses below the protein names. It can be noticed that the mean hydrophobic moments for helices in domain 1 and domain 2 of EPG5 cluster differently: EPG5 domain 1 presents an enrichment of helices towards increased hydrophilicity and high hydrophobic moments that are more similar to those of membrane-remodeling proteins, whereas EPG5 domain 2 shows a clustering resembling more that of karyopherins. (D) Immunoprecipitation with anti-EPG5 or -GFP or anti-RAN antibodies resolved with anti-RAN or anti-EPG5, respectively, and the reciprocal experiment. The same experiment was performed for the XPO1/CRM1-RAN interaction. WCE, whole-cell extract. (E) 3D rendering of HEK293T cells labelled with anti-EPG5 antibody showing its perinuclear distribution (left panel). Nuclear and cytoplasm distribution of EPG5 by immunofluorescence (in HEK293T cells) with EMD antibody (right panel) and by western blot in fibroblasts and LCL of HD (right panel). LMNB and GAPDH were used as reference markers for nuclear and cytoplasmic extracts, respectively.
