Figure 6.
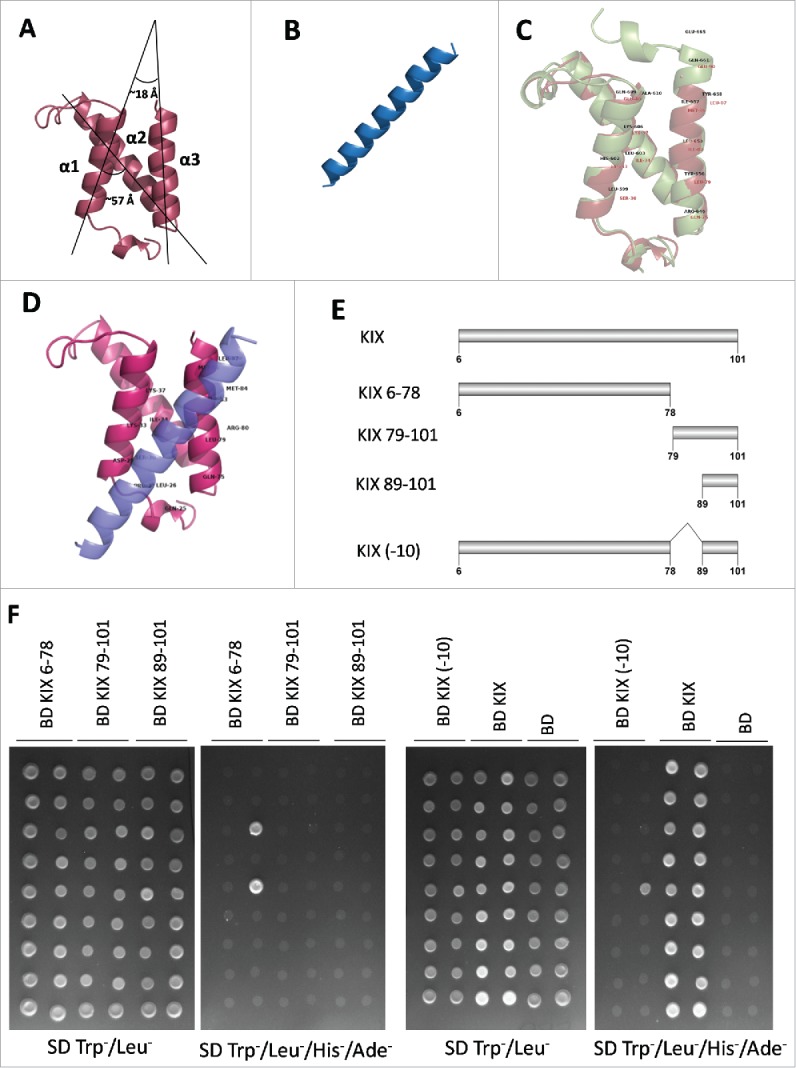
Third helix of AtMED15a KIX is required for protein-protein interactions. (A) Structure of AtMed15a KIX was predicted by homology modelling using mCBP KIX as template. (B) Based on the presence of binding region and 9aaTADs, MYB63 (D176 to D203) peptide was selected for docking studies. (C) Structures of mCBP KIX (green) and AtMed15a KIX (brown) were aligned using PyMOL, Labels show interacting residues. (D) Complex of AtMed15a KIX with MYB63 (D176 to D203), showing interacting residues of AtMed15a KIX. (E) Fragments of AtMed15a KIX were made based on the prediction of interacting residues in α3. (F) Interaction of Y2H positive clones with different fragments of AtMed15a KIX domain, as mentioned in (E), was checked. Cultures of co-transformed yeast were spotted on SD Trp−/Leu− to show proper growth, while on SD Trp−/Leu−/His−/Ade− to score the interactions.
