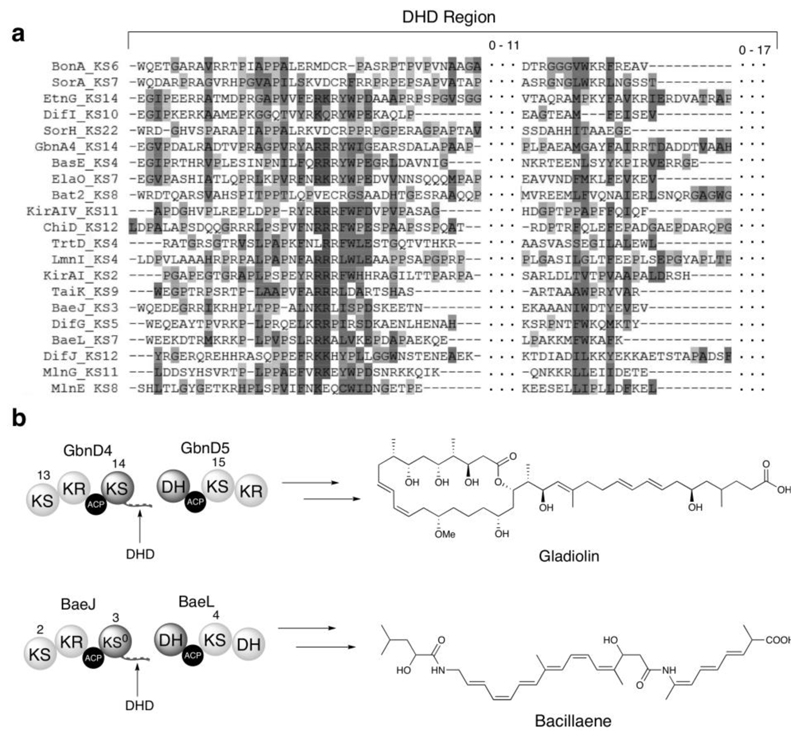
Figure 1. Bioinformatics analysis of DHD domains and examples from the gladiolin and bacillaene PKSs.
(a) Multiple sequence alignment of DHD domains identified by manual inspection of KS/DH boundaries in trans-AT PKSs for which the metabolic product is known. Localized conservation is observed in the N-terminal region, but the C-terminal region is poorly conserved. Dotted sections indicate residues between the conserved regions of the DHD domain, and the number of residues located in these regions is indicated above. Sequences from the following pathways were used: Bae, bacillaene (B. amyloliquefaciens); Bas, basiliskamide (B. laterosporus PE36); Bat, batumin/kalimantacin (P. fluorescens); Dfn, difficidin (B. amyloliquefaciens); Etn, etnangien (S. cellulosum So ce56); Ela, elansolid (Chitinophaga pinensis); Gbn, gladiolin (B. gladioli); Kir, kirromycin (S. collinus); Lnm, leinamycin (S. atroolivaceus); Mln, macrolactin (B. amyloliquefaciens); Mmp, mupirocin (Pseudomonas fluorescens); Sor, sorangicin (Sorangium cellulosum); Tai, thailandamide (B. thailandensis E264); Trt, tartrolon (T. turnerae T7901).
(b) Examples of DHD domains in the gladiolin (top) and bacillaene (bottom) PKSs and structures of the metabolites produced by these assembly lines.