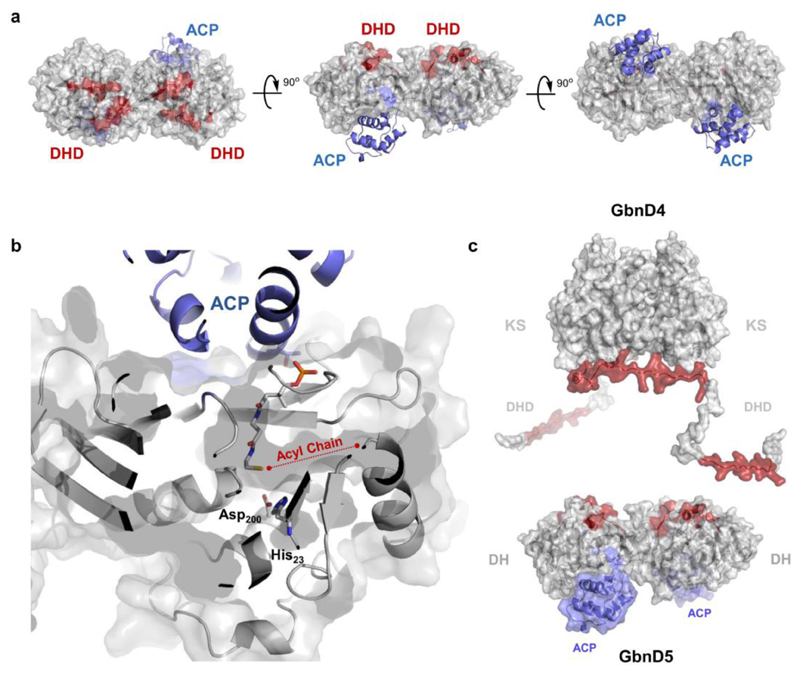
Figure 4. Mapping DHD and ACP domain interaction sites on the DH domain.
(a) Identification of the interaction sites for the GbnD4 DHD domain and GbnD5 ACP domain on the dimeric GbnD5 DH domain using carbene foot-printing. Regions masked by the DHD and ACP domains are highlighted in red and blue, respectively, on a homology model of GbnD5 DH domain (grey). Residue constraints derived from the foot-printing experiments were used to dock the ACP domain (blue) onto the DH domain. Residues used to constrain the docking calculations are as follows; DH: Leu31, Arg61, Leu298, Val299, Asp300; ACP: Leu394, Ala395, Leu396. The model shows adjacent but distinct binding sites for the DHD and ACP domains. These place the GbnD4 KS domain and the GbnD5 ACP domain in close proximity, facilitating efficient communication across the subunit interface.
(b) Predicted location of the ppant arm in the model of the GdnD5 DH-ACP di-domain complex based on the X-ray crystal structure of a crosslinked complex of the ACP and DH from the E. coli fatty acid synthase (PDB accession no: 4KEH). The docking site for the ACP domain is situated near the entrance to the active site of the DH domain. This allows the thiol of the ppant arm to sit adjacent to the catalytic Asp and His residues in the active site of the DH domain. A putative binding pocket for the acyl chain of the substrate sits to the right of the ppant thiol.
(c) Overall model for the interaction between the GbnD4 KS-DHD (red) and GbnD5 DH-ACP (grey/blue) di-domains, both of which form homodimers. The red patches on the surface of the DH and DHD domains indicate mutual interaction sites.