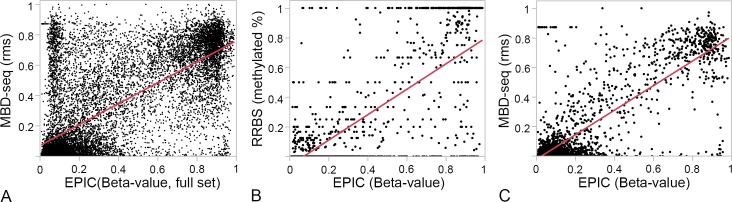
Fig 3. Correlation between MethylationEPIC and high-throughput sequencing data.
Two sequencing datasets were considered for comparison. The MBD-seq was generated in-house from the same samples as the microarray data, and the mouse liver reduced representation bisulfite sequencing (RRBS) data was from a public repository. For each of the 13,665 conserved probes, the 300 bp window around the corresponding CpG was determined and the CpG density-normalized relative methylation score (rms) was estimated for that region from the MBD-seq data. (A) While there was overall significant correlation between the β-values and rms (Pearson’s correlation of 0.70, p < 0.0001), for CpGs with low β-values, the corresponding regions showed rms that cluster close to 0, and for CpG with high β-values, the corresponding rms tended to cluster close to 0.75. (B) For the RRBS data, 2,548 CpGs intersected with the list of 13,665 CpGs interrogated by the conserved EPIC probes. For most CpGs, methylation levels were concordant between the RRBS and EPIC (R = 0.78, p < 0.0001). However, sequence coverage was low for several CpGs in the RRBS data (average total read counts of <7) and such CpGs were associated with estimated methylation fraction of 0 or 1. (C) For the smaller subset of 2,548 CpGs, the β-values showed a more linear correlation with the MBD-seq data (R = 0.84, p < 0.0001).