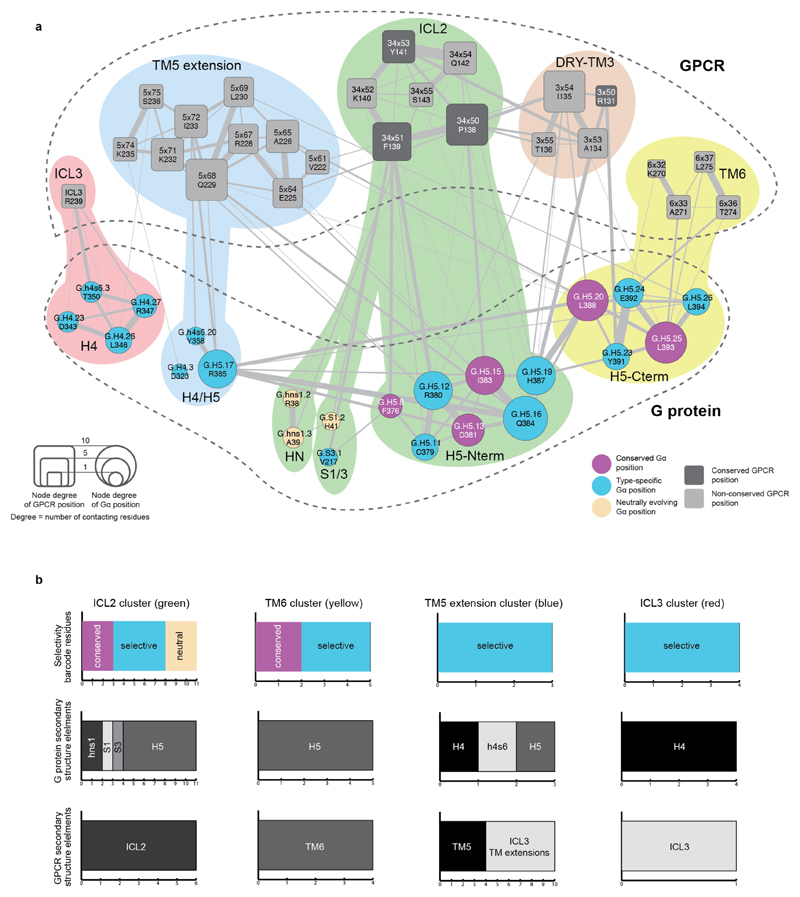
Extended Data Figure 6. Integration of sequence and structure-derived information to understand how GPCRs read the G protein selectivity barcode.
G protein selectivity barcode (Fig. 3d) mapped onto the GPCR-G protein interface clusters obtained using the β2AR-Gs complex structure (Fig. 4; Methods) highlights which regions of the GPCR contact selectivity-determining residues on the G protein. Nodes represent GPCR (rounded squares) and G protein (circles) positions. The edges and their width represent the number of atomic contacts between residues. The size of the nodes is relative to their node degree (number of contacts to other nodes; which is a measure of how central a node is). Residues within the cluster are grouped and coloured differently in the background (red, blue, green, brown and yellow). b, Statistics highlighting the results from integrating the G protein barcode analysis (sequence-based analysis) with the structural clustering analysis (structure-based analysis). The number of residues in Gαs with a particular sequence conservation property in each cluster (i.e. universally conserved, neutrally evolving, selectivity determining position) is shown. The number of residues that map to the different GPCR and G protein secondary structure elements are shown for both GPCR and G protein based on the β2AR-Gs complex structure (PDB: 3sn6).