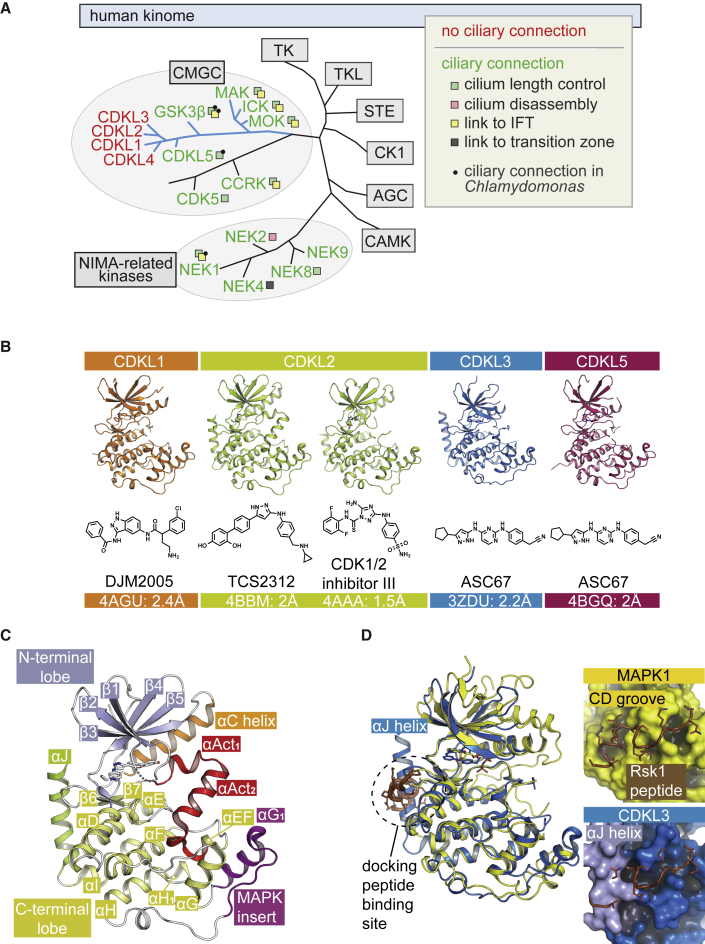
Figure 1.
Unusual Structural Features of the CDKL Kinase Domains
(A) Phylogenetic distribution of NIMA-related kinases (Neks) or CMGC group kinases with known ciliary functions (green), including cilium length control, disassembly, association with intraflagellar transport (IFT), and TZ localization. A branch of the CMGC group (blue) includes several kinases (mammalian MAK, ICK, and MOK; Chlamydomonas GSK3β and CDKL5) that regulate cilium length, several of which also influence IFT. Human CDKL1, CDKL2, CDKL3, and CDKL4 (red) have no previously known links to cilia.
(B) Crystal structures of human CDKL1, CDKL2, CDKL3, and CDKL5 with the indicated inhibitors.
(C) Structural features of the CDKL2-TCS2312 complex, the most complete/ordered CDKL structure.
(D) Structural comparison of CDKL3 with MAPK1 (ERK2; PDB ID: 3TEI). Inset panels show Rsk1 docking peptide bound to MAPK1 and superimposed onto CDKL3.