Abstract
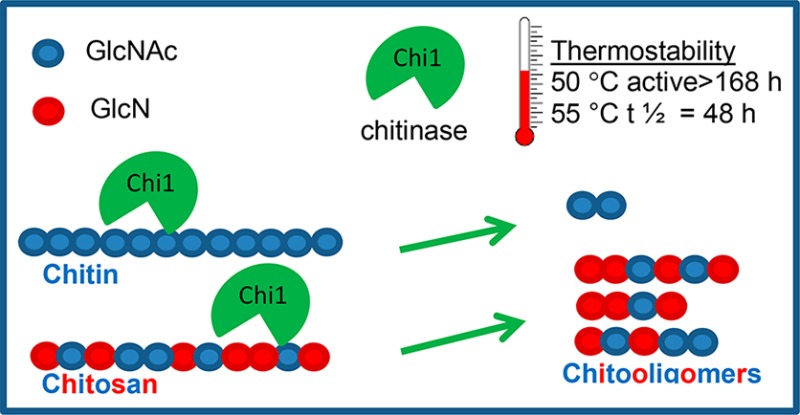
A thermostable Chitinase Chi1 from Myceliophthora thermophila C1 was homologously produced and characterized. Chitinase Chi1 shows high thermostability at 40 °C (>140 h 90% activity), 50 °C (>168 h 90% activity), and 55 °C (half-life 48 h). Chitinase Chi1 has broad substrate specificity and converts chitin, chitosan, modified chitosan, and chitin oligosaccharides. The activity of Chitinase Chi1 is strongly affected by the degree of deacetylation (DDA), molecular weight (Mw), and side chain modification of chitosan. Chitinase Chi1 releases mainly (GlcNAc)2 from insoluble chitin and chito-oligosaccharides with a polymerization degree (DP) ranging from 2 to 12 from chitosan, in a processive way. Chitinase Chi1 shows higher activity toward chitin oligosaccharides (GlcNAc)4–6 than toward (GlcNAc)3 and is inactive for (GlcNAc)2. During hydrolysis, oligosaccharides bind at subsites −2 to +2 in the enzyme’s active site. Chitinase Chi1 can be used for chitin valorisation and for production of chitin- and chito-oligosaccharides at industrial scale.
Keywords: Myceliophthora thermophila C1, chitinase, chitin, chitosan, chito-oligosaccharides
Introduction
Chitin consists of β-(1,4)-linked N-acetyl-d-glucosamine (GlcNAc) units and is one of the most abundant polymers in nature. Chitin is a main component found in shells of crabs, shrimps, and lobsters which are popular types of seafood. Every year approximately 6–8 million tons of shell-waste are produced from the seafood industry globally.1 This waste stream represents a cheap and renewable resource of chitin, which can be used for production of value-added chemicals. Next to crustacean waste, chitin can also be isolated from insects and fungi. Products obtained from chitin and its deacetylated derivative chitosan can be used in medical applications, packaging, food and nutrition, biotechnology, agriculture, and environmental protection.2,3 In recent years, special interest has been paid to water-soluble chito-oligosaccharides, which can act as antimicrobial,4 antitumor, and anti-inflammatory5 agents. Chito-oligosaccharides can be produced by chemical or enzymatic depolymerization of chitin and chitosan. Common procedures for the production of chito-oligosaccharides rely on acid catalysis, which is characterized by a low yield and high environmental impact. The use of enzyme catalysis for depolymerization of chitin and chitosan is a promising alternative to the chemical methods because it allows the production of specific chitin- and chito-oligosaccharides in a controlled way and environmentally friendly process.6 Nevertheless, development of an efficient enzymatic process requires fundamental knowledge of the catalytic mechanisms of enzymes and understanding the interactions with their substrate.
In nature, chitin is degraded by three groups of enzymes: chitinases (EC 3.2.1.14), releasing water-soluble chitin oligosaccharides from chitin, N-acetylglucosaminidases (EC 3.2.1.52), degrading products released by chitinases to monomers,7 and chitin-active lytic polysaccharide monooxygenases (LPMOs; EC 1.14.99.53) that cleave chitin crystalline chains in an oxidative way, yielding a lactone (C1-oxidation) and a ketoaldose (C4-oxidation) product.8−10 The copper-dependent LPMOs act in synergy with chitinases and enhance the accessibility of chitin chains for chitinases and N-acetylglucosaminidases by disrupting the crystal structure of chitin and generation of more soluble polymer chains with increased susceptibility for enzymatic hydrolysis. Based on the amino acid sequence, chitinases have been classified into the glycoside hydrolase (GH) families 18 and 19 and N-acetylglucosaminidase into GH 20 and GH 3, according to the carbohydrate active enzymes classification (CAZy; http://www.cazy.org/).
Chitinases are spread in nature and are involved in physiological processes of bacteria, archea, fungi, animals, and plants.11,12 In recent years special interest has been paid to thermostable chitinases from bacteria and fungi due to their potential application in bioconversion of chitin waste and in the industrial production of chitin oligosaccharides from chitin and chito-oligosaccharides from chitosan. The advantage of thermostable enzymes is that these enzymes do not lose their activity at higher temperatures which are implemented in bioconversion of waste and in industrial processes.6 A number of thermophilic chitinases have been described from bacteria including Chitinophaga, Alcaligenes, Virgibaillus, Massilia, Paenibacillus, Streptomyces, Microbispora, Bacillus, and Brevibacillus.6,13 However, only a few thermophilic fungi have been explored for thermophilic chitinases like Aspergillus fumigatus,14Chaetomium thermophilum,15Gliocladium catenulatum,16Rhizopus oryzae,17Thermoascus aurantiacus vs levisporus,15Thermomyces lanuginosus,19,20 and Trichoderma viridae.21
The thermophilic filamentous fungus Myceliophthora thermophila C1 (previously known as Chrysosporium lucknowense C1; Genencor International B.V., a DuPont company) has been developed for homologous and heterologous protein expression.22M. thermophila C1 has been used before for production of different cell wall degrading enzymes as described by Hinz and co-workers.23,24 We reported before that M. thermophila C1 produces an endochitinase entitled Chitinase Chi1.22 Dua et al.25 published recently an exochitinase rMtChit from M. thermophila BJA produced from the same gene sequence as Chitinase Chi1 by recombinant expression in Pichia pastoris; however, characteristics of exochitinase rMtChit differed significantly from our Chitinase Chi1. Since Chitinase Chi1 from M. thermophila C1 might have potential application in bioconversion of chitin waste sources and in industrial production of chitin and chito-oligosaccharides, a full characterization of the Chitinase Chi1 and understanding of the interactions of Chitinase Chi1 with its substrates is important. Here, we describe the production and detailed characterization of Chitinase Chi1 with focus on thermostability, catalytic properties, and mode of action on chitin, chitosan, and chitin and chito-oligosaccharides.
Materials and Methods
Chemicals
Chitin azure, chitin from shrimp shells, glycol chitosan, Schiff’s reagent, 4-nitrophenyl-N-acetylglucosamine (GlcNAc-pNP), 4-nitrophenyl-N,N′-diacetyl-β-d-chitobioside ((GlcNAc)2-pNP), and 4-nitrophenyl-β-d-N,N′,N″-triacetylchitotriose ((GlcNAc)3-pNP), were obtained from Sigma-Aldrich (St. Louis, USA). Oxidized chitosan (Mw 100 kDa, DDA 84%, degree of oxidation 5%, containing C6-aldehyde and carboxyl groups in a ratio of 20:1) was produced at Wageningen Food & Biobased Research (Wageningen, The Netherlands). Hydroxypropyl-chitosan was a kind gift from Nippon Suisan (Japan). Chitin oligosaccharides (GlcNAc)2–6 were obtained from Megazyme (Co. Wicklow, Ireland). Chitosans were purchased from Heppe Medical Chitosan GmbH (Halle, Germany) and Nippon Suisan Kaisha LTD (Tokyo, Japan). The deacetylation degree (DDA in %) and molecular weight (Mw in kDa) are chitosan 88 DDA/3000 and chitosan 90 DDA/100 (Nippon Suisan Kaisha LTD), chitosan 77 DDA/600, chitosan 78 DDA/600, chitosan 91 DDA/600 and chitosan 94 DDA/600 (Heppe Medical Chitosan GmbH). All other chemicals were of the highest purity available.
Swollen Chitin Preparation
Swollen chitin was prepared according to Monreal and Reese26 with some modifications. Chitin from shrimp shells (1 g) was stirred in 25 mL of 85% (v/v) phosphoric acid and left at room temperature for 20 h. Subsequently it was precipitated by pouring the gelatinous mixture into an excess of ice-cold water. The swollen chitin was separated by centrifugation at 3000g and washed with demineralized water up to pH 6.0.
Overexpression of Chi1 Gene in M. thermophila C1 and 2 L-Scale Fermentation for Production of Chitinase Chi1
Overexpression of Chitinase Chi1 in M. thermophila C1 and the 2 L-scale fermentation for production of Chitinase Chi1 have been carried out based on the procedures previously reported: (i) the DNA sequence of the gene encoding (Chi1) for Chitinase Chi1 (GenBank accession number HI550986) was described in a patent,27 (ii) the homologous overexpression of Chitinase Chi1 in M. thermophila C1 (previously known as C. lucknowense C1) was described in detail by Visser et al.22 and (iii) the preparation of a monocomponent strain of M. thermophila C1 producing Chitinase Chi1 and the detailed conditions of the fed-batch fermentation resulting in high production level of Chitinase Chi1 (7.5 g/L) were also described by Visser et al.22 In short, the Chi1 gene was amplified from genomic M. thermophila C1 DNA and cloned into a M. thermophila C1 expression vector. The expression cassette containing the chi1 promoter, gene and the terminator obtained from the vector was transformed into a low protease/(hemi)cellulase free M. thermophila C1-expression host. Ninety six transformants were grown in a microtiter plate28 and screened for Chitinase Chi1 production levels in the culture broth using chitin azure as substrate. The transformant showing the highest level of chitinase activity was selected for fed-batch fermentation to produce Chitinase Chi1 at 2-L scale. The strain was grown aerobically in 2 L fermenters in mineral medium, containing glucose as carbon source, ammonium sulfate as nitrogen source and trace elements for the essential salts. The enzyme was produced at pH 6.0 and 32 °C.27 The supernatant containing Chitinase Chi1 was centrifuged at 20 000g for 20 min to remove the biomass and concentrated (4 fold) using a 5 kDa PES membrane (Vivacell 70, Sartorius). The crude enzyme extract was subsequently dialyzed against 10 mM potassium phosphate buffer pH 6.0 and freeze-dried to obtain the crude enzyme preparation. Freeze-drying was used to prevent microbial decay of enzyme preparation and to avoid the use of preservatives.
Sequence Analysis of Chitinase Chi1
Nucleotide and deduced amino acid sequences were analyzed using Clone Manager software. BLAST analysis of the deduced amino acid sequence of Chitinase Chi1 was performed at the NCBI server (https://blast.ncbi.nlm.nih.gov/Blast.cgi). Analysis for conserved domains was performed using Conserved Domain Search and Conserved Domain Database (containing information from databases including Pfam, SMART, COG, PRK, TIGRFAM)29 at the NCBI server (https://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi). The signal peptide was analyzed at the SignalP 4.0 server (http://www.cbs.dtu.dk/services/SignalP/) and the theoretical isoelectric point (pI) was calculated with Compute pI/Mw tool on ExPASy server (https://web.expasy.org/compute_pi/). Potential N-linked glycosylation sites and potential O-linked glycosylation sites were predicted by NetOGlyc 4.0 Server and NetNGlyc 1.0 Server (http://www.cbs.dtu.dk/services/). To generate a 3D model of Chitinase Chi1 the deduced amino acid sequence of Chitinase Chi1 was submitted to the Phyre2 web portal for protein modeling.30 The Phyre2 generated the protein model on the basis of its closest template.
Purification of Chitinase
The freeze-dried crude enzyme preparation containing 142 mg of total protein was dissolved in 50 mL of 0.05 M Bis-Tris buffer pH 7.0 and purified by anion exchange chromatography using a HiPrep DEAE FF 16/10 column (GE Healthcare Bio-Science AB, Uppsala, Sweden) and an ÄKTA pure system (GE Healthcare Bio-Science AB, Uppsala, Sweden). The column was equilibrated with five column volumes (CV) 0.05 M Bis-Tris buffer pH 7.0 (buffer A). A sample of 50 mL was loaded onto the column and eluted using 0.05 M Bis-Tris buffer pH 7.0 followed by elution with 1 M NaCl in 0.05 M Bis-Tris buffer pH 7.0 (buffer B) as follows: 20% buffer B for 10 CV and 45% buffer B for 10 CV with a flow rate of 5 mL min–1. Fractions were collected and screened for chitinase and N-acetylglucosaminidase activity. The fraction containing (10 mL) the highest chitinase activity was subjected to size exclusion chromatography and loaded onto a HiLoad 16/600 Superdex 75 pg column (GE Healthcare Bio-Science AB, Uppsala, Sweden). Proteins were eluted isocratic with 0.05 M Bis-Tris buffer pH 7.0 containing 0.15 M NaCl with a flow rate of 0.5 mL min–1. The absorbance was measured at 280 nm.
Enzyme Assays and Protein Concentration
During the purification process the activities of chitinase and N-acetylglucosaminidase were measured. Chitinase activity was determined using colloidal chitin azure31 as substrate. The enzyme solution (0.05 mL) was incubated with 0.95 mL 5% (w/v) colloidal chitin azure in 50 mM Bis-Tris buffer pH 7.0, and the mixture was incubated at 50 °C for 30 min. After incubation, the reaction was terminated by heating at 96 °C for 5 min, to inactivate the enzyme. The reaction mixture was centrifuged at 20 000g for 5 min, and the absorbance of the supernatant was measured at 560 nm. An enzyme-free mixture was used as negative control, and each reported value was the average of duplicate tests. One enzyme unit was defined as a change in the absorbance of 0.01 min–1.32
For N-acetylglucosaminidase activity, the reaction mixture contained 0.09 mL of 2 mM GlcNAc-pNP in 0.1 M citrate-phosphate buffer pH 4 and 0.01 mL enzyme solution. After 10 min incubation in a microtiter plate at 50 °C, 0.2 mL of 0.25 M Tris/HCl buffer pH 8.8 was added to the mixtures and the absorbance at 405 nm was measured using a Tecan Safire plate reader (Grodig, Austria). One enzyme activity was defined as the amount of enzyme that liberated 1 μmol of pNP per minute. The protein concentration was determined using the bicinchoninic acid assay (BCA) according to the recommendation of the supplier (Pierce) with bovine serum albumin as standard.
Sodium Dodecyl Sulfate-Polyacrylamide Gel Electrophoresis (SDS-PAGE) Identification of Glycosylated Proteins and Isoelectric Point Determination
Sodium dodecyl sulfate-polyacrylamide (10% (w/v)) gel electrophoresis (SDS-PAGE) was performed as described by Laemmli.33 A NuPAGE Novex System (ThermoFisher Scientific, Bleiswijk, The Netherlands) with 10% (w/v) Bis-Tris gels was used. Prior to electrophoresis, all samples were heated for 10 min at 70 °C in NuPAGE LDS sample buffer with NuPAGE sample reducing agent, according to the instructions of the manufacturer. Gels were stained with SimplyBlue SafeStain according to the recommendation of the supplier (ThermoFisher Scientific). For detection of glycosylated proteins, the SDS-PAGE gel was stained using periodic acid-Schiff staining (PAS).34 The gel was incubated subsequently for 1 h in 12.5% (w/v) trichloroacetic acid, 1 h in 1% (v/v) periodic acid/3% (v/v) acetic acid, 1 h in 15% (v/v) acetic acid (replaced every 10 min), and 1 h at 4 °C in the dark in Schiff’s reagent (Sigma-Aldrich, Zwijndrecht, The Netherlands). Hereafter, the gel was washed two times for 5 min in 0.5% (w/v) sodium bisulfite and destained in 7% (v/v) acetic acid. The isoelectric point (pI) of Chitinase Chi1 was estimated by isoelectric focusing (IEF) using PhastGel IEF on a Pharmacia LKB Phast System (Pharmacia Biotech, Uppsala, Sweden) with a broad protein calibration kit (pH 3–10, GE Healthcare) as standard. Proteins were stained with Coomassie blue R-2.
Mass Spectrometry
The molecular weight of Chitinase Chi1 was determined by matrix assisted laser-desorption time-of-flight mass spectrometry (MALDI-TOF-MS). Samples were prepared by the dried droplet method on a 600 μm AnchorChip target (Bruker), using 5 mg mL–1 2,5-dihydroxyacetophenone, 1.5 mg mL–1 diammonium hydrogen citrate, 25% (v/v) ethanol, and 3% (v/v) trifluoroacetic acid as matrix. Spectra were derived from ten 500-shots (1000 Hz) acquisitions taken at nonoverlapping locations across the sample. Measurements were made in the positive linear mode, with ion source 1, 25.0 kV; ion source 2, 23.3 kV; lens, 6.5 kV; pulsed ion extraction, 680 ns. Protein Calibration Standard II (Bruker) was used for external calibration.
Purity and Identity of Chitinase Chi1
To evaluate the identity and purity of Chitinase Chi1, a sample containing the purified Chitinase Chi1 was sent to The Scripps Research Institute, Proteomics Core (Jupiter, FL, U.S.A.) for proteolytic digestion and HPLC-ESI-MS/MS analysis.
Influence of Temperature and pH on Activity and Stability of Chitinase Chi1
Influence of temperature on activity of Chitinase Chi1 was analyzed with swollen chitin as substrate in the range of 30–80 °C. The enzyme solution (0.05 mL) containing 1.6 μM Chitinase Chi1 was incubated with 0.95 mL of 1% (w/v) swollen chitin in 0.1 M citrate-phosphate-borate buffer pH 6.0 at 50 °C while mixing at 800 rpm for 30 min. The reaction was terminated by heating at 96 °C for 5 min. The reaction mixture was centrifuged at 20 000g for 10 min. The produced reducing sugars in the supernatant were measured using the p-hydroxybenzoic acid hydrazide (PAHBAH) assay.35 An enzyme-free mixture was used as negative control, and each reported value was the average of duplicate tests. N-Acetyl-d-glucosamine was used as a standard, and one enzyme unit (U) was defined as the amount of enzyme that liberated 1 μmol reducing sugar per minute. The thermostability was determined by preincubating the purified Chitinase Chi1 (1.6 μM) at pH 6.0 (0.1 M citrate-phosphate-borate buffer) at various temperatures (40–60 °C) for different time intervals up to 168 h. The influence of pH on activity of Chitinase Chi1 was determined by incubating Chitinase Chi1 (1.6 μM) at different pH levels (3.0–9.0) in 0.1 M citrate-phosphate-borate buffer using swollen chitin as substrate.
Determination of Kinetic Parameters
The Km, Vmax, and kcat values for swollen chitin and chitosan 90 DDA/100 kDa were calculated with GraphPad Prism software (GraphPad Software, U.S.A.). The reducing sugars produced in the supernatant were measured using the p-hydroxybenzoic acid hydrazide (PAHBAH) assay.35
Depolymerization of Chitosans with Different Mw and DDA
To elucidate the influence of Mw and DDA of chitosans on Chitinase Chi1 activity a wide range of different chitosans were tested including: glycol chitosan, hydroxypropyl chitosan, oxidized chitosan, and chitosans with different DDA and Mw. Chitosans were used in a concentration of 0.1% (w/v) with 40.7 nM of purified Chitinase Chi1 in 1 mL of 0.05 M sodium phosphate buffer pH 6.0. The mixture was incubated at 50 °C while mixing at 800 rpm for 15 min. After incubation, the enzyme activity was terminated by heating at 96 °C for 5 min. The reaction mixture was centrifuged at 20 000g for 5 min. The reducing sugars produced in the supernatant were measured using the p-hydroxybenzoic acid hydrazide (PAHBAH) assay.35
Hydrolysis of Swollen Chitin and Chitosan
For the enzymatic hydrolysis of swollen chitin and chitosan 90 DDA/100 the reaction mixtures containing 1 mL of 0.45% (w/v) substrate in 0.05 M sodium phosphate buffer pH 6.0 with 100 nM purified Chitinase Chi1 were incubated at 50 °C while mixing at 800 rpm. Aliquots were taken at different time intervals, and the hydrolysis products were analyzed by high-performance anion-exchange chromatography (HPAEC) and MALDI-TOF-MS.
Hydrolysis of Chitin Oligosaccharides and pNP-Substrates
Hydrolysis of chitin oligosaccharides (GlcNAc)2–6 and pNP-substrates was followed in time. Incubations were performed with 25 nM purified Chitinase Chi1 in 0.5 mL reaction volume containing 2 mM substrate (GlcNAc)2–6, 1 mM (GlcNAc)2-pNP or (GlcNAc)3-pNP, and 50 mM sodium phosphate buffer pH 6.0. Samples were incubated at 50 °C and aliquots of 60 μL were taken at different time intervals. The reaction was terminated by heating at 96 °C for 5 min, and the hydrolysis products were analyzed by HPAEC.
HPAEC
An ICS-3000 ion chromatography HPLC system equipped with a CarboPac PA-1 column (2 × 250 mm) in combination with a CarboPac PA-guard column (2 × 25 mm) at 22 °C and a pulsed electrochemical detector (PAD) in pulsed amperometric detection mode (Dionex) at 30 °C was used. A flow rate of 0.25 mL min–1 was used, and the column was equilibrated with water. The following gradient was used: 0–25 min H2O, 25–65 min at 0–0.045 M NaOH, 65–70 min at 0.045 M NaOH-1 M sodium acetate in 0.1 M NaOH, 70–75 min at 1 M sodium acetate in 0.1 M NaOH, 75–75.1 min 1 M sodium acetate in 0.1 M NaOH-0.1 M NaOH, 75.1–80 min 0.1 M NaOH, and 80–95 min H2O. Post column addition was used for increasing the PAD signal by 0.5 M NaOH at a flow rate of 0.15 mL min–1.
Identification of Chitin and Chito-oligosaccharides by MALDI-TOF-MS
MALDI-TOF-MS was performed on a Bruker UltraFlextreme (Bruker Daltonics) in reflective mode, and positive ions were examined. The instrument was calibrated for positive ions with a mixture of maltodextrin standards with known molecular masses. Samples were diluted in the matrix solution containing 10 mg mL–1 2,5-dihydroxybenzoic acid in 50% (v/v) acetonitrile. For analysis, 1 μL of the mixture was transferred to the target plate and dried under a stream of dry air. The lowest laser intensity needed to obtain a good quality spectrum was applied, and 10 times 50 laser shots randomly obtained from the sample were accumulated.
Results
Sequence Analysis of Chitinase Chi1
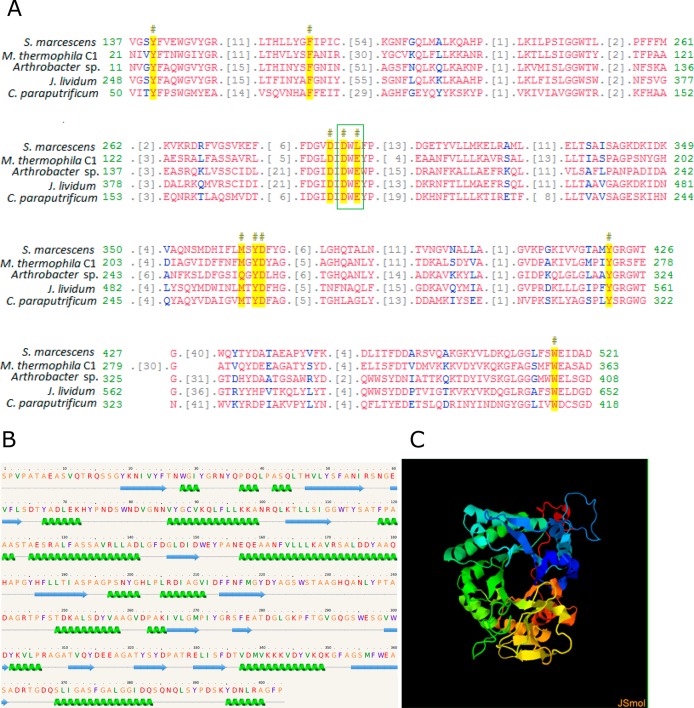
The putative gene chi1 encoding for Chitinase Chi1 had an ORF of 1281-bp that encoded a protein with 426 amino acids, in which a signal peptide is predicted that consists of 23 amino acids in the N-terminal region of the protein. The deduced molecular weight of Chitinase Chi1 was 43.8 kDa, and a theoretical pI was at pH 4.95. Four potential O-linked glycosylation sites, one potential N-linked glycosylation site, and 47 phosphorylation sites were found in the sequence. Multiple sequence alignment of the active site of the deduced protein sequence of Chitinase Chi1 (Figure 1A) with Chitinase A from Serratia marcescens (1NH6_A), Chitinase B from Arthrobacter sp. TAD20 (1KFW_A), Janthinobacterium lividum (AAA83223), and Chitinase from Clostridium paraputrificum (BAD12045) revealed the presence of the conserved glycoside GH 18 domain in Chitinase Chi1. In total 10 conserved amino acids and one conserved active-site motif consisting of aspartate (D) and glutamate (E) residues forming the D-X-E motif were found in Chitinase Chi1. Modeling of the secondary structure of Chitinase Chi1 revealed that Chitinase Chi1 is composed of 16 α-helixes and 14 β-sheets (Figure 1B). 3D modeling of Chitinase Chi1 was based on similarity with Chitinase from the fungus Clonostachys rosea belonging to GH 18 (template PDB entry: 3G6MA; Figure 1C). Using this sequence 386 amino acids residues (equal to 96% of the whole amino acid sequence) have been modeled with 100% confidence by the single highest scoring template. This modeling revealed the (β/α)8 barrel fold (TIM) of Chitinase Chi1.
Figure 1.
Sequence analysis of Chitinase Chi1 from Myceliophthora thermophila C1. A. Multiple sequence alignment of active site of Chitinase Chi1 from Myceliophthora thermophila C1 with active site of Chitinase A from Serratia marcescens (1NH6_A), Chitinase B from Arthrobacter sp. TAD20 (1KFW_A), Janthinobacterium lividum (AAA83223), and Chitinase from Clostridium paraputrificum (BAD12045). Conserved residues are colored in yellow and marked with (#) sign. Conserved D-X-E motif is shown in the green box. Chitinase Chi1 shares 10 amino acids with conserved hydrolases family 18 (GH 18) domain. Analysis was performed with Conserved Domain Search and Conserved Domain Database. B. Secondary structure of Chitinase Chi1 predicted with Phyre2. C. 3D modeling of Chitinase Chi1 was performed with Phyre2 software with Chitinase from Clonostachys rosea belonging to GH 18 family (PDB entry 3G6MA) used as template.
Purification of Chitinase Chi1
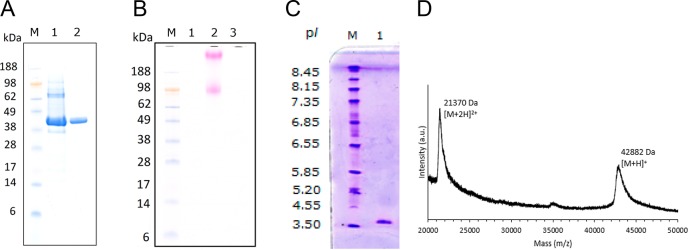
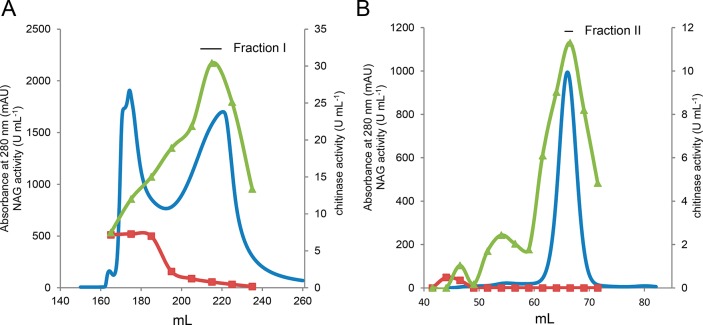
The gene chi1 encoding for Chitinase Chi1 was successfully cloned into the M. thermophila C1-expression host. The transformant with the highest production was used for the production of high amounts of Chitinase Chi1, (7.5 g/L), in 2-L fermentation. From the culture broth, 15 g of protein containing about 60% Chitinase Chi1 (based on SDS-PAGE, Figure 3A) was obtained. The crude enzyme preparation was further subjected to a two-step purification process using anion exchange and size exclusion chromatography. The first purification step (Figure 2A) resulted in the separation of the extract in two main protein peaks of which the peak eluting first showed mainly N-acetylglucosaminidase activity and the second peak chitinase activity. However, in the second peak some N-acetylglucosaminidase activity was detected. Therefore, in order to remove this activity, fraction I from the first purification step was subjected to size exclusion chromatography using a Superdex 75 (Figure 2B). This step enabled a clear separation between Chitinase Chi1 and the remaining N-acetylglucosaminidase. Chitinase Chi1 was successfully purified to homogeneity as shown on SDS-PAGE as one single band (Figure 3A) and was confirmed by HPLC-ESI-MS/MS analysis of the proteolytic digest (Figure S1, Supporting Information) that identified only peptides originating from Chitinase Chi1. Staining with periodic acid-Schiff (PAS) did not detect any glycosylation of Chitinase Chi1 (Figure 3B). The specific activity of the purified Chitinase Chi1 was 3.5 U mg–1 for colloidal chitin azure (Table 1).
Figure 3.
Analysis of purified Chitinase Chi1 from Myceliophthora thermophila C1. A. molecular weight of Chitinase Chi1 determined by SDS-PAGE under reducing denaturing conditions. Lanes: M, standard protein molecular weight markers; 1, crude enzyme preparation of Chitinase Chi1; 2, purified Chitinase Chi1. B. Protein staining with periodic acid (PAS). Lanes: M, standard protein molecular weight markers; 1, purified Chitinase Chi1; 2, yeast invertase; 3, bovine serum albumin. C. Isoelectric focusing (IEF). Lanes: M, pI marker; 1, purified Chitinase Chi1. D. Molecular weight of Chitinase Chi1 determined by MALDI-TOF-MS.
Figure 2.
Purification of Chitinase Chi1 from Myceliophthora thermophila C1 by ion exchange chromatography on DEAE-FF Sepharose (A) and size exclusion chromatography on Superdex 75 (B). Proteins (blue line) were detected at 280 nm. Chitinase activity (green line) was measured with chitin azure at pH 6.0 and 50 °C. Activity of N-acetylglucosaminidase (red line)was assayed with GlcNAc-pNP at pH 4.0 and 50 °C.
Table 1. Characterization of Chitinase-Containing Fractions Obtained during Purification of Chitinase Chi1 from Myceliophthora thermophila C1.
| purification step | volume (mL) | total activity (U) | total protein (mg) | specific activity (U mg–1)a |
|---|---|---|---|---|
| crude extract | 50 | 373 | 142 | 2.6 |
| anion exchange chromatography fraction I | 10 | 126 | 51 | 2.5 |
| size exclusion chromatography fraction II | 2.5 | 23 | 6.6 | 3.5 |
Specific activity was assayed with 5% (w/v) colloidal chitin azure at 50 °C and pH 7.0 and was calculated per mg protein.
Molecular Weight and Isoelectric Point of Chitinase Chi1
Chitinase Chi1 is a monomeric polypeptide, and the molecular weight predicted from the protein sequence is 43.8 kDa. The molecular weight of the purified Chitinase Chi1 was measured by MALDI-TOF-MS and was shown to be 42.9 kDa (Figure 3D). The MALDI-TOF-MS spectrum of Chitinase Chi1 showed an intense signal of the single charged protein [M + H]+ at m/z 42 882 and the signal of the double-charged protein [M+2H]2+ at m/z 21 370. The SDS-PAGE revealed a molecular weight of 43 kDa (Figure 3A). The isoelectric point of Chitinase Chi1 was found to be 3.95 (Figure 3C).
Influence of pH and Temperature on Activity and Stability of Chitinase Chi1
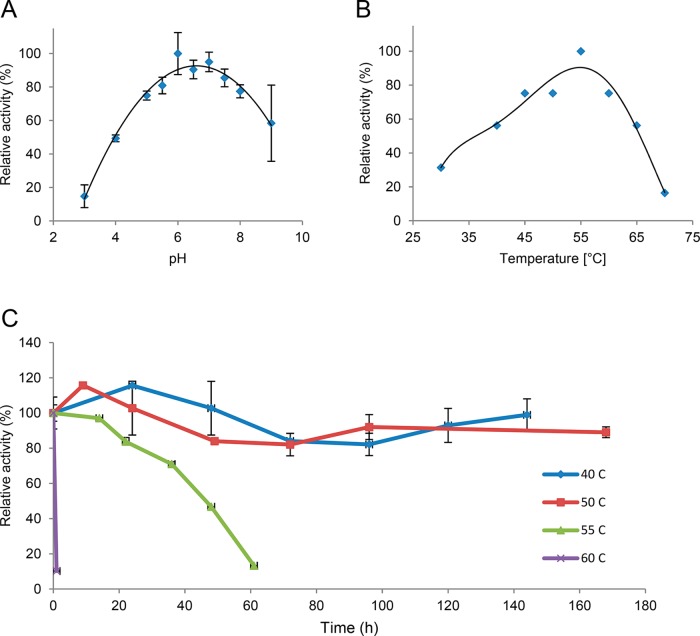
Chitinase Chi1 was active at pH 3.0 to 9.0 (Figure 4A) and exhibited the highest activity at pH 6.0. Chitinase Chi1 showed activity from 30 to 70 °C with the highest activity at 55 °C (Figure 4B), and it was remarkably stable at 40 °C (>140 h 90% activity) and 50 °C (>168 h 90% activity; Figure 4C). At 55 °C the enzyme reached its half-life after 48 h. Incubation at 60 °C resulted in fast inactivation of the enzyme, with a loss of 90% of the initial catalytic activity after 1 h incubation.
Figure 4.
Enzyme characteristics of Chitinase Chi1: A. Enzyme activities at various pH (pH 3.0–9.0) were measured at 50 °C in 0.1 M citrate-phosphate-borate buffer. B. Enzyme activities at various temperatures (30–80 °C) were measured at pH 6.0 in 0.1 M citrate-phosphate-borate buffer. C. Thermostability was measured by incubating Chitinase Chi1 at various temperatures (40–60 °C), and the residual activities were assayed at 50 °C. Reactions were performed with 1.6 μM purified Chitinase Chi1 and 1% (w/v) swollen chitin in 0.1 M citrate-phosphate-borate buffer for 30 min. The error bars represent the range of duplicate experiments.
Kinetic Parameters for Chitinase Chi1
Kinetic parameters of Chitinase Chi1 were determined for swollen chitin and for chitosan 90 DDA/100. Activities were determined based on reducing sugars released during the reaction.
For swollen chitin the specific activity was 1.4 ± 0.2 U mg–1, Vmax was 12.2 ± 0.5 μM min–1, Km was 2.0 ± 0.2 mg mL–1, and kcat was 0.11 ± 0.0 s–1.
For chitosan the specific activity was 10.0 ± 0.6 U mg–1, Vmax was 194 ± 21.4 μM min–1, Km was 0.9 ± 0.3 mg mL–1, and kcat was 1.9 ± 0.2 s–1.
Influence of Molecular Weight and Degree of Deacetylation of Chitosan on Chitinase Chi1 Activity
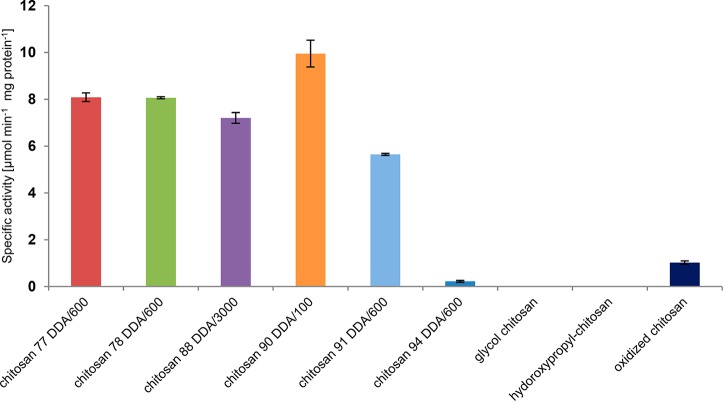
To evaluate the influence of the Mw (100, 600, and 3000 kDa) and DDA (77, 78, 88, 90, and 94) of chitosan on Chitinase Chi1 activity, a range of chitosans was tested (Figure 5). In case of chitosans with the same Mw (600 kDa), Chitinase Chi1 showed decreased activity when DDA was increased. The highest activity was measured for chitosan with the lowest Mw (chitosan 90 DDA/100). For chitosan with similar DDA but different Mw (chitosan 88 DDA/3000 and chitosan 90 DDA/100), Chitinase Chi1 showed higher activity for lower Mw chitosan. Chitinase Chi1 degraded partially oxidized chitosan (i.e., chitosan with random oxidation at C6 positions) but showed 10% of activity as measured for the untreated parent chitosan (chitosan 90 DDA/100). Chitinase Chi1 was not able to degrade glycol chitosan and hydroxypropyl-chitosan, which are both fully deacetylated.
Figure 5.
Specific activity of Chitinase Chi1 on different types of chitosans. Chitosans were used in a concentration of 0.1% (w/v) with 40.7 nM of purified Chitinase Chi1 in 1 mL of 0.05 M sodium phosphate buffer pH 6.0. The reaction mixtures were incubated at 50 °C, for 15 min. The reducing sugars produced in the supernatant were measured using the p-hydroxybenzoic acid hydrazide (PAHBAH) assay. The results represent the average of duplicate experiments.
Degradation of Chitin and Chitosan by Chitinase Chi1
Degradation of chitin incubated with Chitinase Chi1 was followed in time, and products released were analyzed by HPAEC. The main products formed were (GlcNAc)2 next to a small amount of (GlcNAc)3 and GlcNAc (Figure 6). After the first 30 min the rate of (GlcNAc)2 release gradually decreased in time. The concentration of (GlcNAc)3 and GlcNAc increased gradually up to 30 min of incubation and leveled off hereafter. At 90 min incubation the concentration of (GlcNAc)3 started to decrease, when degradation into (GlcNAc)2 and GlcNAc became the predominant reaction. The ratio (GlcNAc)2 to GlcNAc at the end of the reaction was equal to 11. Chitin conversion expressed as the amount of (GlcNAc)2 produced was 3.1% after 6 h.
Figure 6.
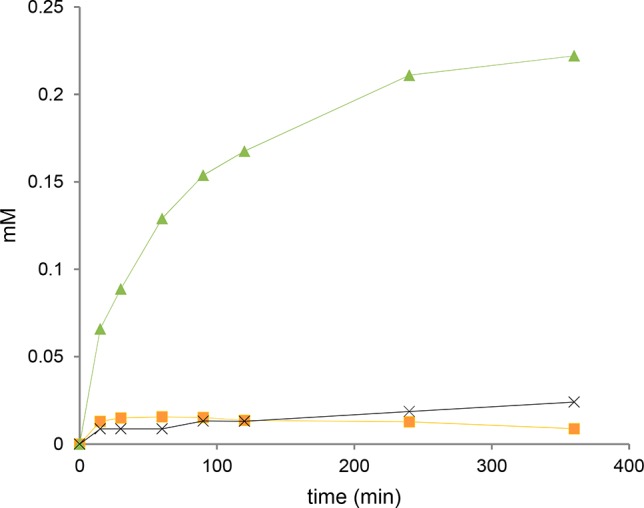
Release of chitin oligosaccharides during swollen chitin hydrolysis by Chitinase Chi1. Swollen chitin (0.45% (w/v)) was incubated with purified Chitinase Chi1 (100 nM) at 50 °C in 1 mL of 0.05 M sodium phosphate buffer pH 6.0. Aliquots were taken at different time intervals, and the hydrolysis products were analyzed by HPAEC. Detected products were GlcNAc (cross), (GlcNAc)2 (triangle), and (GlcNAc)3 (square). Soft lines are only drawn as visual aids. The error bars represent the range of duplicate experiments.
Products released from chitosan 90 DDA/100 were analyzed by MALDI-TOF-MS. Chito-oligosaccharides were detected as potassium and/or sodium adducts and are summarized in Table S1 (Supporting Information). Chitinase Chi1 was able to release a whole spectrum of hetero-oligosaccharides consisting of GlcNAc and GlcN units with a polymerization degree (DP) ranging from 2 to 12. The chito-oligosaccharide composition of the reaction mixture changed over time (Figure S2, Supporting Information). At the early stages of the reaction, a large diversity of chito-oligosaccharides were formed, containing 2–6 GlcNAc units and 1–9 GlcN residues in different combinations and with a ratio GlcNAc/GlcN spanning the range between 0.5 and 4 (Table 1S and Figure S2, Supporting Information). During the reaction, chito-oligosaccharides with more than three GlcNAc residues might be further degraded by Chitinase Chi1, and new GlcN-enriched chito-oligosaccharides containing up to 10 GlcN residues and one or two GlcNAc residues are formed that accumulate in time (Table 1S and Figure S3, Supporting Information). Fully acetylated DP 2 ((GlcNAc)2, 447.2 m/z [M + Na]+) was identified from the early stages of the reaction and during the entire incubation. Fully acetylated DP 3 (650.3 m/z [M + Na]+) was detected only at 15 min incubation time, indicating that it was subsequently degraded by the enzyme. Accumulation of the heterologous dimer (DP 2) composed of GlcNAc and GlcN (405.2 m/z [M + Na]+) at longer reaction times, in the slow phase of the reaction, may indicate the ability of Chitinase Chi1 to cleave glycosidic linkages between GlcN and GlcNAc moieties in the chitosan chain but only when GlcNAc is positioned in −1 subsite of the enzyme active site.
Degradation of Chitin Oligosaccharides and pNP-Substrates by Chitinase Chi1
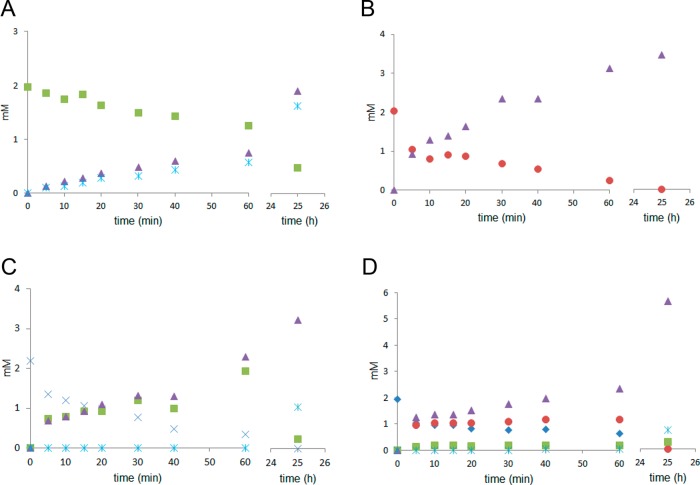
In order to study the binding preferences and to determine the shortest possible substrate for Chitinase Chi1 the hydrolysis of chito-oligosaccharides (GlcNAc)2–6 was followed in time and the hydrolysis products were analyzed by HPAEC (Figure 7). (GlcNAc)2 was not hydrolyzed by Chitinase Chi1 (data not shown), whereas (GlcNAc)3 was cleaved to (GlcNAc)2 and GlcNAc (Figure 7A). (GlcNAc)4 was split only to (GlcNAc)2 (Figure 7B). (GlcNAc)5 was degraded to (GlcNAc)2 and (GlcNAc)3; however, after 25 h (GlcNAc)3 was degraded to GlcNAc and (GlcNAc)2 (Figure 7C). Depolymerization of (GlcNAc)6 resulted in the initial release of (GlcNAc)4 and (GlcNAc)2 and small amounts of (GlcNAc)3 (Figure 7D). The released (GlcNAc)4 was further degraded to (GlcNAc)2, and (GlcNAc)3 was cleaved to (GlcNAc)2 and GlcNAc. The calculated rates for the degradation of chitin oligosaccharides were 0.18 mM min–1 for (GlcNAc)6, 0.17 mM min–1 for (GlcNAc)5, 0.20 mM min–1 for (GlcNAc)4, and 0.02 mM min–1 for (GlcNAc)3.
Figure 7.
Hydrolysis of chitin oligosaccharides (GlcNAc)3 (A), (GlcNAc)4 (B), (GlcNAc)5 (C), and (GlcNAc)6 (D) by Chitinase Chi1. Chitin oligosaccharides (2 mM) in 0.5 mL of 0.05 M sodium phosphate buffer pH 6.0 were incubated with purified Chitinase Chi1 (25 nM). Aliquots were taken at different time intervals, and the hydrolysis products were analyzed by HPAEC. Detected products were GlcNAc (star), (GlcNAc)2 (triangle), (GlcNAc)3 (square), (GlcNAc)4 (circle), (GlcNAc)5 (cross), and (GlcNAc)6 (diamond). Experimental work performed in duplicates and the standard deviation was less than 5%.
Among pNP-labeled chitin oligosaccharides, activity of Chitinase Chi1 was detected for (GlcNAc)3-pNP and (GlcNAc)2-pNP with the highest specific activity for (GlcNAc)2-pNP (Figure S3A, Supporting Information). No activity was found for GlcNAc-pNP, which is in agreement that no activity was detected for (GlcNAc)2.
To investigate the reaction mechanism in more detail, degradation of labeled chitin oligosaccharides (GlcNAc)3-pNP and (GlcNAc)2-pNP was followed in time by HPAEC (Figure S3B, Supporting Information). Hydrolysis of (GlcNAc)3-pNP yielded predominantly GlcNAc-pNP and (GlcNAc)2 (about 90% end product) and low amounts of (GlcNAc)3 (about 10% end product). In time the released (GlcNAc)3 was degraded further to (GlcNAc)2 and GlcNAc. Products detected from the hydrolysis of (GlcNAc)2-pNP were (GlcNAc)2, GlcNAc-pNP, and small amounts of (GlcNAc)3 (Figure S4B, Supporting Information). The substrate used in this experiment did not contain (GlcNAc)3 (Figure S4C, Supporting Information). Production of (GlcNAc)3 can be explained by the transglycosylation reaction catalyzed by Chitinase Chi1, and it is in agreement with results reported for Chitinase MBP-CfcA from Aspergillus niger.36
Discussion
Analysis of the amino acid sequence of Chitinase Chi1 confirmed that Chitinase Chi1 is a real glycoside hydrolase from GH 18 family, which contains characteristic for this family D-X-E motif. The 3D modeled structure revealed that Chitinase Chi1 has (β/α)8 barrel fold (TIM) which is another characteristic for chitinases from GH 18.
Chitinase Chi1 was purified to homogeneity as confirmed by MALDI-TOF-MS and HPLC-MS/MS analysis of the proteolytic digests of the purified enzyme. The increase in specific activity after each purification step was not very extensive, and this might be due to the additive activity of the accompanying N-acetylglucosaminidase that was detected in the crude enzyme extract and which was still present in the fractions obtained after the first purification step using anion exchange chromatography. The ability of N-acetylglucosaminidase to act on the amorphous parts of chitin was reported for β-N-acetylhexosaminidase (LeHex20A) by Konno and co-workers.37 The second step of purification on size exclusion chromatography enabled a clear separation between Chitinase Chi1 and the remaining N-acetylglucosaminidase, resulting in the isolation of a pure enzyme.
Molecular weight of Chitinase Chi1 measured with MALDI-TOF-MS was 43 kDa, which was different from a molecular weight calculated from the deduced protein sequence of 43.8 kDa. This difference shows that full-length Chitinase Chi1 undergoes post-translational proteolytic modification in the host M. thermophila C1. Detection of peptides from C-terminus end of Chitinase Chi1 with HPLC-MS/MS indicates that proteolytic removal takes place from the N-terminus end of Chitinase Chi1. According to the calculated molecular weight, approximately 9 amino acids might be removed. Proteolytic processing has been previously described for chitinases from other microorganisms including Chitinase ChiC from Serratia marcescens(38) and chitinases from Streptomyces olivaceoviridis(39) and Janthinobacterium lividum.40 Recently, Dua et al.25 reported the exochitinase rMtChit obtained by heterologous expression of the same gene sequence obtained from M. thermophila BJA in Pichia pastoris. This latter protein has a molecular weight of 48 kDa. The difference in molecular weight between Chitinase Chi1 and rMtChit can be explained by the fact that different cloning approaches and different hosts were used for enzyme production; that is, Chitinase Chi1 was expressed homologously in M. thermophila, while exochitinase rMtChit was expressed heterologously in the yeast P. pastoris. Furthermore, Chitinase Chi1 and exochitinase rMtChit differ clearly in the extent of glycosylation. Staining with PAS confirmed that Chitinase Chi1 was not or hardly glycosylated because no magenta color formation was detected for Chitinase Chi1 at a concentration of 1 mg mL–1. In contrast, exochitinase rMtChit gave an intense magenta color, indicating glycosylation, as reported by Dua et al.25
Chitinase Chi1 was found to have a pH optimum at pH 6.0 and a pI of 3.98, while the calculated pI from the amino acid sequence was 4.95. This difference between the theoretical pI predicted from the primary structure and the experimentally determined pI is common, since the pI of proteins is affected by several factors, including the solvent accessibility of amino acids. Some charged amino acids could be shielded by the folded structure of the enzyme and may not be exposed to the solvent, changing therefore the observed pI.
The highest activity of Chitinase Chi1 was detected at 55 °C which is in agreement with other thermophilic chitinases (Table 2). Chitinase Chi1 showed excellent thermostability at 50 °C (>168 h, 90% activity) and at 55 °C (t1/2 = 48 h). Reported thermostable fungal chitinases have also thermostability up to 50 °C, but all are less stable in time than Chitinase Chi1 (Table 2). For example, the exochitinase rMtChit retained only 70% of its activity when exposed to 45 °C for 5 h and showed a t1/2 = 113 min at 65 °C. During incubation at 50 °C (1 h) Chitinase from T. lanuginosus retained 71% of its activity, whereas other Chitinase from T. lanuginosus was able to preserve about 70% of the enzyme activity after 6 h at 50 °C. In contrast, another Chitinase from T. lanuginosus SY2 was 100% active for 1 h when incubating at 50 °C. Therefore, it can be concluded that Chitinase Chi1 can be classified as thermostable chitinase which shows excellent thermostability among other chitinases from thermophilic fungi.
Table 2. Properties of Chitinases from Thermophilic Fungi.
| organism | molecular weight (kDa) | pH optimum | temp. optimum (°C) | temp. range (°C) | thermostability (as % activity retained activity) | ref |
|---|---|---|---|---|---|---|
| Aspergillus fumigatus YJ-407 | 46 | 5 | 50–60 | 45–60 | 100% activity after 1 h at 45 °C | (14) |
| ∼70% activity after 1 h at 55 °C | ||||||
| ∼20% activity after 1 h at 60 °C | ||||||
| Chaetomium thermophilum | 47 | 5.5 | 60 | 40–70 | 100% activity after 1 h at 50 °C | (15) |
| ∼90% activity after 1 h at 60 °C | ||||||
| Gliocladium catenulatum | 51 | 5.0–6.0 | 60 | 20–70 | 40% activity after 20 min at 50 °C | (16) |
| Rhizopus oryzae | 5.5–6.0 | 60 | 50–70 | 100% activity after 30 min at 50 °C | (17) | |
| ∼40% activity after 30 min at 60 °C | ||||||
| Myceliophthora thermophila BJA | 48 | 5.0 | 55 | 30–70 | 70% activity after 5h at 45 °C | (25) |
| ∼50% activity after 113 min at 65 °C | ||||||
| ∼50% activity after 48 min 75 °C | ||||||
| Thermoascus aurantiacus vs levisporus | 48 | 8.0 | 50 | 40–60 | ∼90% activity after 1 h at 50 °C | (15) |
| ≈30% activity after 1 h at 60 °C | ||||||
| Thermomyces lanuginosus | 44.1 | 5.0 | 50 | 30–60 | 50% activity at 50 °C after 630 min | (18) |
| 56% activity at 60 °C after 30 min | ||||||
| Thermomyces lanuginosus | 36.6 | 4.0 | 40 | 30–60 | 50% activity at 40 °C after 577.5 min | (18) |
| 71% activity at 50 °C after 60 min | ||||||
| Thermomyces lanuginosus SY2 | 48 | 55 | 55 | 50–60 | 100% activity after 1 h at 50 °C | (19) |
| ∼80% activity after 1 h at 60 °C | ||||||
| Thermomyces lanuginosus | 42 | 60 | 60 | 30–70 | 70% activity after 6 h at 50 °C | (20) |
| Trichoderma viride | 28 | 3.5 | 55–60 | ∼30% activity after 3 h at 50 °C | (22) | |
| ∼10% activity after 3 h at 60 °C | ||||||
| Myceliophthora thermophila C1 | 43 | 6.0 | 55 | 30–70 | ∼90% activity after >168 h at 50 °C | this work |
| 50% activity after 48 h at 55 °C | ||||||
| 10% activity after 10 min at 60 °C |
Besides activity on chitin, Chitinase Chi1 showed also activity on the deacetylated chitin derivative, chitosan. Solubilized chitosan was more efficiently degraded by the enzyme than swollen chitin. This is in agreement with previous studies reporting that chitinases from GH 18 are able to cleave the glycosidic linkage of not only GlcNAc–GlcNAc but also GlcNAc–GlcN present in chitosan as long as a GlcNAc residue is bound at the −1 subsite.42 Furthermore, higher activity on chitosan than on chitin indicates that substrate accessibility is an important parameter influencing chitinase activity as it was also observed for bacterial chitinase from Ralstonia sp.(41)
The activity of Chitinase Chi1 was strongly affected by DDA, Mw, and presence of side groups (i.e., aldehyde and carboxyl) at the chitosan chain. In general, chitosan with a lower DDA (77 DDA) was degraded more efficiently than chitosan with high DDA (94 DDA). This result confirmed that Chitinase Chi1 is a real GH 18 enzyme, which is dependent on the acetyl group of GlcNAc for catalysis. Thus, a decrease in the number of GlcNAc moieties present in the chitosan chain will result in less productive binding sites of Chitinase Chi1. Chitosan with low Mw (100 kDa) was degraded more efficiently than chitosan with high Mw (3000 kDa) and the same DDA. This result denotes, that chitosan with higher Mw, which also shows higher viscosity, is less accessible for the enzyme than the chitosan with a low Mw. Fully deacetylated modified chitosans with pending aliphatic side groups, such as glycol chitosan and hydroxypropyl-chitosan, were not degraded at all by the enzyme, showing that Chitinase Chi1 is a real chitinase, which is not able to cleave GlcN–GlcN bonds. Although steric hindrance due to side chains cannot be excluded, the results clearly suggest that the Chitinase Chi1 activity depends on the presence and the number of acetyl groups. It was shown for enzymes from GH 18 that the carbonyl oxygen from GlcNAc moiety act as a nucleophile during catalytic reaction of GH 18 enzymes. Similar activity on chitosan was reported for Chitinase from Streptomyces griseus.42
Chitinase Chi1 released mainly (GlcNAc)2 from chitin. Release of dimers was reported for other chitinases from GH 18 family.7,43 In contrast, exochitinase rMtChit released only monomers from chitin.25 Although the amino acid sequence of Chitinase Chi1 and exochitinase rMtChit should be the same, differences in expression host and glycosylation influence the activity and mode of action of Chitinase Chi1 and exochitinase rMtChit.
Next to (GlcNAc)2, Chitinase Chi1 released a small amount of GlcNAc and (GlcNAc)3, with a ratio of (GlcNAc)2/GlcNAc equal to 11. This ratio is commonly used for a rough assessment of enzyme processivity.44,45 Thus, Chitinase Chi1 may be considered as a processive chitinase. A decrease in the rate of chitin hydrolysis, which was observed after 30 min reaction is most likely due to the fact that Chitinase Chi1 enriched the recalcitrant regions of the substrate. It was previously stated that the activity of processive enzymes tends to decrease as the substrate is consumed and when the enzymes reach regions that hinder processive binding.45
The release of (GlcNAc)2 in higher molar amount than other chitin oligosaccharides from chitin was observed for processive and nonprocessive enzymes degrading recalcitrant polysaccharides, like for processive chitinases ChiA and ChiB and nonprocessive ChiC from S. marcescens.43 Therefore, results obtained for Chitinase Chi1 with natural substrates indicate its processivity, but this conclusion is not indisputable. An important feature of processive exoacting chitinases is the so-called α + β insertion domain that forms one “wall” of the substrate binding cleft, which were found in ChiA and ChiB.46,9 This domain was not found in Chitinase Chi1. However, the active site of Chitinase Chi1 is aligned with 60 amino acid residues, which may be important for interactions with the substrate and promote processivity of the enzyme. Aromatic residues in the active site of processive enzymes were shown to interact with the substrate during the processive mode of action.45
Processivity has been studied for other chitinases and also for cellulases.47 It was shown that processive enzymes slide with their active site on the single-polymer chain and stay closely associated with the substrate between subsequent hydrolytic reactions. During the processive degradation of chitin the enzymes release mainly dimers because the successive sugar units in the polymer are rotated by 180° and sliding of such polymers through the enzyme’s active site will result in the productive binding only after every second sugar moiety. Rotation of the sugar units is particularly important for chitinases from family GH 18, since these enzymes require a correctly positioned N-acetyl group in their −1 subsite.43
It may be concluded that Chitinase Chi1 is an endochitinase with a high degree of processivity. It was stated that both endo- and exomechanisms can be combined with processive action46 and that the most important difference between chitinases may be related to the ability of the enzymes to act in processive or nonprocessive ways, rather than to their binding preferences (endo- or exomanner).9
Chitinase Chi1 was able to release a broad spectrum of chitin- and chito-oligosaccharides (DP2-DP12) from chitosan with 90% DDA. The chito-oligosaccharide composition of the reaction mixture changed over time, indicating that release and further degradation of some chito-oligosaccharides might be simultaneously catalyzed by Chitinase Chi1, as it was shown in the experiment with chitin oligosaccharides (GlcNAc)2–6. The composition of chito-oligosaccharides and the accumulation of a (GlcNAc, GlcN) dimer at longer reaction time may indicate the ability of Chitinase Chi1 to cleave glycosidic linkages between GlcN and GlcNAc moieties in the chitosan chain, as it was reported for the Chitinase G from Streptomyces coelicolor A3(2) from bacterial family GH19 Chitinase48 and other bacterial chitinases.49 Release of longer chito-oligosaccharides may indicate that, similarly to chitin degradation, Chitinase Chi1 can degrade chitosan in a processive way. Chitinases ChiA and ChiB from S. marcescens(50) were shown to degrade chitosan in a processive way. In the case of chitosan, processive enzymes stay attached to the substrate after productive (with a correctly positioned N-acetyl group in the sugar unit) or nonproductive (with lack of a correctly positioned N-acetyl group in the sugar unit) initial binding to the substrate. Binding of the substrate will be followed by sliding of the substrate through the active site cleft by two sugar units at the time, until a new productive complex will be formed and an enzymatic reaction occurs.43
Chitinase Chi1 degraded chitin oligosaccharides with DP ≥ 3. The absence of activity on (GlcNAc)2 ruled out the possibility that Chitinase Chi1 is an N-acetylglucosaminidase. All initial released chitin oligosaccharides with DP ≥ 3 were subject to further hydrolysis that yielded (GlcNAc)2 and GlcNAc as final products. Chitinase Chi1 showed increasing activities with increasing DP of chitin oligosaccharides. These data indicate that Chitinase Chi1 has a multisubsite binding cleft and positioning of chitin oligosaccharides with DP ≥ 4 is more efficient than with DP 3, resulting in about 10 times faster conversion for longer-chain chitin oligosaccharides than for shorter ones. Additionally, experiments conducted with pNP-(GlcNAc)3 and pNP-(GlcNAc)2 confirmed that oligosaccharides bind to Chitinase Chi1 at subsites −2 to +2 in the active site during hydrolysis.
Overall, we showed here that homologous expressed Chitinase Chi1 releases mainly dimers from chitin and might use a processive mechanism. Depolymerization of chitosan resulted in the production of a wide range of chito-oligosaccharides. Chitin and chito-oligosaccharides are an emerging class of bioactive ingredients with potential biomedical, cosmetic, and pharmaceutic applications. The need for green and biocompatible technologies for the production of chitin and chito-oligosaccharides reveals new perspectives for the application of biocatalysts. With its remarkable thermostability and activity in a wide range of pH, Chitinase Chi1 is a promising biocatalyst for bioconversion of chitin waste sources and production of chitin and chito-oligosaccharides from both chitin and chitosan at industrial scale.
Glossary
Abbreviations Used
- DDA
degree of deacetylation
- DP
polymerization degree
- GlcNAc
N-acetyl-d-glucosamine
- (GlcNAc)2
diacetyl-chitobiose
- (GlcNAc)3
triacetyl-chitotriose
- (GlcNAc)4
tetraacetyl-chitotetraose
- (GlcNAc)5
pentaacetyl-chitopentaose
- (GlcNAc)6
hexaacetyl-chitohexaose
- GlcNAc-pNP
4-nitrophenyl–N-acetylglucosamine
- (GlcNAc)2-pNP
4-nitrophenyl-N,N′-diacetyl-β-d-chitobioside
- (GlcNAc)3-pNP
4-nitrophenyl-β-d-N,N′,N″-triacetylchitotriose
- LPMO
lytic polysaccharide monooxygenase
- GH 18
glycoside hydrolase family 18
- GH 20
glycoside hydrolase family 20
- GH 3
glycoside hydrolase family 3
- Mw
molecular weight
- SDS-PAGE
sodium dodecyl sulfate-polyacrylamide gel electrophoresis
- pI
isolectric point
- PAHBAH
p-hydroxybenzoic acid hydrazide
- MALDI-TOF-MS
matrix assisted laser-desorption time-of-flight mass spectrometry
- HPAEC
high performance anion-exchange chromatography
Supporting Information Available
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acs.jafc.7b04032.
Protein oligomers detected after proteolytic digestion of Chitinase Chi1 with HPLC-ESI-MS/MS analysis (Figure S1). Spectra from matrix assisted laser-desorption time-of-flight mass spectrometry (MALDI-TOF-MS) obtained for chito-oligosaccharides released by Chitinase Chi1 from chitosan 90 DDA/100 and their molecular mass (Figure S2 and Table S1). Specific activity of Chitinase Chi1 for GlcNAc-pNP, (GlcNAc)2-pNP, and (GlcNAc)3-pNP (A) and mode of action of Chitinase Chi1 on (GlcNAc)3-pNP and (GlcNAc)2-pNP (B) (Figure S3). HPAEC elution profiles of standards of chito-oligosaccharides (GlcNAc)1–3 and GlcNAc-pNP (A) and reaction products released after 60 min incubation of (GlcNAc)2-pNP with purifed Chitinase Chi1 (B) (Figure S4). (PDF)
This research received funding from The Netherlands Organisation for Scientific Research (NWO) in the framework of the TASC Technology Area BIOMASS.
The authors declare no competing financial interest.
Supplementary Material
References
- Gao X.; Chen X.; Zhang J.; Guo W.; Jin F.; Yan N. Transformation of chitin and waste shrimp shells into acetic acid and pyrrole. ACS Sustainable Chem. Eng. 2016, 4, 3912–3920. 10.1021/acssuschemeng.6b00767. [DOI] [Google Scholar]
- Kardas I.; Struszczyk M. H.; Kucharska M.; Van den Broek L. A. M.; Van Dam J. E. G.; Ciechańska D.. Chitin and chitosan as functional biopolymers for industrial applications. In The European Polysaccharide Network of Excellence (EPNOE); Navard P., Ed.; Springer: Vienna, 2012; pp 329–373. [Google Scholar]
- Van den Broek L. A. M.; Knoop R. J. I.; Kappen F. H. J.; Boeriu C. G. Chitosan films and blends for packaging material. Carbohydr. Polym. 2015, 116, 237–242. 10.1016/j.carbpol.2014.07.039. [DOI] [PubMed] [Google Scholar]
- No H. K.; Young Park N.; Ho Lee S.; Meyers S. P. Antibacterial activity of chitosans and chitosan oligomers with different molecular weights. Int. J. Food Microbiol. 2002, 74, 65–72. 10.1016/S0168-1605(01)00717-6. [DOI] [PubMed] [Google Scholar]
- Azuma K.; Osaki T.; Minami S.; Okamoto Y. Anticancer and anti-inflammatory properties of chitin and chitosan oligosaccharides. J. Funct. Biomater. 2015, 6, 33–49. 10.3390/jfb6010033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meena S.; Gothwal R. K.; Mohan M. K.; Ghosh P. Production and purification of a hyperthermostable Chitinase from Brevibacillus formosus BISR-1 isolated from the Great Indian Desert soils. Extremophiles 2014, 18, 451–462. 10.1007/s00792-014-0630-4. [DOI] [PubMed] [Google Scholar]
- Ike M.; Nagamatsu K.; Shioya A.; Nogawa M.; Ogasawara W.; Okada H.; Morikawa Y. Purification, characterization, and gene cloning of 46 kDa Chitinase (Chi46) from Trichoderma reesei PC-3–7 and its expression in Escherichia coli. Appl. Microbiol. Biotechnol. 2006, 71, 294–303. 10.1007/s00253-005-0171-y. [DOI] [PubMed] [Google Scholar]
- Vaaje-Kolstad G.; Westereng B.; Horn S. J.; Liu Z. L.; Zhai H.; Sørlie M.; Eijsink V. G. H. An oxidative enzyme boosting the enzymatic conversion of recalcitrant polysaccharides. Science 2010, 330, 219–222. 10.1126/science.1192231. [DOI] [PubMed] [Google Scholar]
- Vaaje-Kolstad G.; Horn S. J.; Sørlie M.; Eijsink V. G. H. The chitinolytic machinery of Serratia marcescens – a model system for enzymatic degradation of recalcitrant polysaccharides. FEBS J. 2013, 280, 3028–3049. 10.1111/febs.12181. [DOI] [PubMed] [Google Scholar]
- Forsberg Z.; Nelson C. E.; Dalhus B.; Mekasha S.; Loose J. S. M.; Crouch L. I.; Røhr Å.K.; Gardner J. G.; Eijsink V. G. H.; Vaaje-Kolstad G. Structural and functional analysis of a lytic polysaccharide monooxygenase important for efficient utilization of chitin in Cellvibrio japonicus. J. Biol. Chem. 2016, 291, 7300–7312. 10.1074/jbc.M115.700161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adrangi S.; Faramarzi M. A.; Shahverdi A. R.; Sepehrizadeh Z. Purification and characterization of two extracellular endochitinases from Massilia timonae. Carbohydr. Res. 2010, 345, 402–407. 10.1016/j.carres.2009.11.015. [DOI] [PubMed] [Google Scholar]
- Hartl L.; Zach S.; Seidl-Seiboth V. Fungal chitinases: diversity, mechanistic properties and biotechnological potential. Appl. Microbiol. Biotechnol. 2012, 93, 533–543. 10.1007/s00253-011-3723-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nawani N. N.; Kapadnis B. P.; Das A. D.; Rao A. S.; Mahajan S. K. Purification and characterization of a thermophilic and acidophilic chitinase from Microbispora sp. V2. J. Appl. Microbiol. 2002, 93, 965–975. 10.1046/j.1365-2672.2002.01766.x. [DOI] [PubMed] [Google Scholar]
- Xia G.; Jin C.; Zhou J.; Yang S.; Zhang S.; Jin C. A novel chitinase having a unique mode of action from Aspergillus fumigatus YJ-407. Eur. J. Biochem. 2001, 268, 4079–4085. 10.1046/j.1432-1327.2001.02323.x. [DOI] [PubMed] [Google Scholar]
- Li A.-N.; Yu K.; Liu H.-Q.; Zhang J.; Li H.; Li D.-C. Two novel thermostable Chitinase genes from thermophilic fungi: Cloning, expression and characterization. Bioresour. Technol. 2010, 101, 5546–5551. 10.1016/j.biortech.2010.02.058. [DOI] [PubMed] [Google Scholar]
- Ma G.-Z.; Gao H.-L.; Zhang Y.-H.; Li S.-D.; Xie B.-Y.; Wu S.-J. Purification and characterization of Chitinase from Gliocladium catenulatum strain HL-1–1. Afr. J. Microbiol. Res. 2012, 6, 4377–4383. 10.5897/AJMR12.605. [DOI] [Google Scholar]
- Chen W.-M.; Chen C.-S.; Jiang S.-T. Purification and characterization of an extracellular Chitinase from Rhizopus Oryzae. J. Mar Sci. Technol. 2013, 21, 361–366. [Google Scholar]
- Zhang M.; Puri A. K.; Govender A.; Wang Z.; Singh S.; Permaul K. The multi-chitinolytic enzyme system of the compost-dwelling thermophilic fungus Thermomyces lanuginosus. Process Biochem. 2015, 50, 237–244. 10.1016/j.procbio.2014.11.008. [DOI] [Google Scholar]
- Guo R.-F.; Shi B.-S.; Li D.-C.; Ma W.; Wei Q. Purification and characterization of a novel thermostable chitinase from Thermomyces lanuginosus SY2 and cloning of its encoding gene. Agric. Sci. China 2008, 7, 1458–1465. 10.1016/S1671-2927(08)60403-4. [DOI] [Google Scholar]
- Prasad M.; Palanivelu P. Overexpression of a chitinase gene from the thermophilic fungus, Thermomyces lanuginosus in saccharomyces cerevisiae and characterization of the recombinant chitinase. J. Microb. Biochem. Technol. 2012, 4, 86–91. 10.4172/1948-5948.1000076. [DOI] [Google Scholar]
- Omumasaba C. A.; Yoshida N.; Ogawa K. Purification and characterization of a Chitinase from Trichoderma viride. J. Gen. Appl. Microbiol. 2001, 47, 53–61. 10.2323/jgam.47.53. [DOI] [PubMed] [Google Scholar]
- Visser H.; Joosten V.; Punt P. J.; Gusakov A.; Olson P. T.; Joosten R.; Bartels J.; Visser J.; Sinitsyn A.; Emalfarb M.; Verdoes J.; Wery J. Development of a mature fungal technology and production platform for industrial enzymes based on a Myceliophthora thermophila isolate, previously known as Chrysosporium luknowense C1. Ind. Biotechnol. 2011, 7, 214–223. 10.1089/ind.2011.7.214. [DOI] [Google Scholar]
- Hinz S. W. A.; Pouvreau L.; Joosten R.; Bartels J.; Jonathan M. C.; Wery J.; Schols H. A. Hemicellulase production in Chrysosporium lucknowense C1. J. Cereal Sci. 2009, 50, 318–323. 10.1016/j.jcs.2009.07.005. [DOI] [Google Scholar]
- Kühnel S.; Hinz S. W. A.; Pouvreau L.; Wery J.; Schols H. A.; Gruppen H. Chrysosporium lucknowense arabinohydrolases effectively degrade sugar beet arabinan. Bioresour. Technol. 2010, 101, 8300–8307. 10.1016/j.biortech.2010.05.070. [DOI] [PubMed] [Google Scholar]
- Dua A.; Joshi S.; Satyanarayana T. Recombinant Exochitinase of the Thermophilic Mould Myceliopthora thermophila BJA: Characteristics and Utility in Generating N-acetyl Glucosamine and in Biocontrol of Phytopathogenic Fungi. Biotechnol. Prog. 2017, 33, 70–80. 10.1002/btpr.2370. [DOI] [PubMed] [Google Scholar]
- Monreal J.; Reese E. T. The Chitinase of Serratia marcescens. Can. J. Microbiol. 1969, 15, 689–696. 10.1139/m69-122. [DOI] [PubMed] [Google Scholar]
- Verdoes J. C.; Punt P. J.; Burlingame R. P.; Pynnonen C. M.; Olson P. T.; Wery J.; Visser J. H.; Emalfarb M. A. V. J.. New fungal production system. Int. Patent WO/2010/107303, 2010.
- Verdoes J. C.; Punt P. J.; Burlingame R.; Bartels J.; Dijk R.; Slump E.; Meens M.; Joosten R.; Emalfarb M. A dedicated vector for efficient library construction and high throughput screening in the hyphal fungus Chrysosporium lucknowense. Ind. Biotechnol. 2007, 3, 48–57. 10.1089/ind.2007.3.048. [DOI] [Google Scholar]
- Marchlerbauer A.; Derbyshire M. K.; Gonzales N. R.; Lu S.; Chitsaz F.; Geer L. Y.; Geer R. C.; He J.; Gwadz M.; Hurwitz D. I. CDD: NCBI’s conserved domain database. Nucleic Acids Res. 2015, 43, 222–226. 10.1093/nar/gku1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelley L. A.; Mezulis S.; Yates C. M.; Wass M. N.; Sternberg M. J. E. The Phyre2 web portal for protein modeling, prediction and analysis. Nat. Protoc. 2015, 10, 845–858. 10.1038/nprot.2015.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen C.-R.; Chen Y.-S.; Yang C.-J.; Chen J.-K.; Liu C.-L. Colloid chitin azure is a dispersible, low-cost substrate for Chitinase measurements in a sensitive, fast, reproducible assay. J. Biomol. Screening 2010, 15, 213–217. 10.1177/1087057109355057. [DOI] [PubMed] [Google Scholar]
- Gómez Ramírez M.; Rojas Avelizapa L. I.; Rojas Avelizapa N. G.; Cruz Camarillo R. Colloidal chitin stained with Remazol Brilliant Blue R®, a useful substrate to select chitinolytic microorganisms and to evaluate chitinases. J. Microbiol. Methods 2004, 56, 213–219. 10.1016/j.mimet.2003.10.011. [DOI] [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 1970, 227, 680–685. 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Zacharius R. M.; Zell T. E.; Morrison J. H.; Woodlock J. J. Glycoprotein staining following electrophoresis on acrylamide gels. Anal. Biochem. 1969, 30, 148–152. 10.1016/0003-2697(69)90383-2. [DOI] [PubMed] [Google Scholar]
- Lever M. Colorimetric and fluorometric carbohydrate determination with p-hydroxybenzoic acid hydrazide. Biochem. Med. 1973, 7, 274–281. 10.1016/0006-2944(73)90083-5. [DOI] [PubMed] [Google Scholar]
- Van Munster J. M.; Dobruchowska J. M.; Veloo R.; Dijkhuizen L.; van der Maarel M. J. E. C. Characterization of the starvation-induced Chitinase CfcA and α-1,3-glucanase AgnB of Aspergillus niger. Appl. Microbiol. Biotechnol. 2015, 99, 2209–2223. 10.1007/s00253-014-6062-3. [DOI] [PubMed] [Google Scholar]
- Konno N.; Takahashi H.; Nakajima M.; Takeda T.; Sakamoto Y. Characterization of β-N-acetylhexosaminidase (LeHex20A), a member of glycoside hydrolase family 20, from Lentinula edodes (shiitake mushroom). AMB Express 2012, 2, 29. 10.1186/2191-0855-2-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gal S. W.; Choi J. Y.; Kim C. Y.; Cheong Y. H.; Choi Y. J.; Lee S. Y.; Bahk J. D.; Cho M. J. Cloning of the 52-kDa Chitinase gene from Serratia marcescens KCTC2172 and its proteolytic cleavage into an active 35-kDa enzyme. FEMS Microbiol. Lett. 1998, 160, 151–158. 10.1111/j.1574-6968.1998.tb12905.x. [DOI] [PubMed] [Google Scholar]
- Romaguera A.; Menge U.; Breves R.; Diekmann H. Chitinases of Streptomyces olivaceoviridis and signicance of processing for multiplicity. J. Bacteriol. 1992, 174, 3450–3454. 10.1128/jb.174.11.3450-3454.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gleave A. P.; Taylor R. K.; Morris B. A.; Greenwood D. R. Cloning and sequencing of a gene encoding the 69-kDa extracellular Chitinase of Janthinobacterium lividum. FEMS Microbiol. Lett. 1995, 131, 279–288. 10.1111/j.1574-6968.1995.tb07788.x. [DOI] [PubMed] [Google Scholar]
- Sutrisno A.; Ueda M.; Abe Y.; Nakazawa M.; Miyatake K. A Chitinase with high activity toward partially N-acetylated chitosan from a new, moderately thermophilic, chitin-degrading bacterium, Ralstonia sp. A-471. Appl. Microbiol. Biotechnol. 2004, 63, 398–406. 10.1007/s00253-003-1351-2. [DOI] [PubMed] [Google Scholar]
- Ohtakara A.; Matsunaga H.; Mitsutomi M. Action pattern of Streptomyces griseus chitinase on partially N-acetylated chitosan. Agric. Biol. Chem. 1990, 54, 3191–3199. 10.1271/bbb1961.54.3191. [DOI] [PubMed] [Google Scholar]
- Horn S. J.; Sørbotten A.; Synstad B.; Sikorski P.; Sørlie M.; Vårum K. M.; Eijsink V. G. H. Endo/exo mechanism and processivity of family 18 chitinases produced by Serratia marcescens. FEBS J. 2006, 273, 491–503. 10.1111/j.1742-4658.2005.05079.x. [DOI] [PubMed] [Google Scholar]
- Teeri T. T.; Koivula A.; Linder M.; Wohlfahrt G.; Divne C.; Jones T. A. Trichoderma reesei cellobiohydrolases: why so efficient on crystalline cellulose?. Biochem. Soc. Trans. 1998, 26, 173–177. 10.1042/bst0260173. [DOI] [PubMed] [Google Scholar]
- Hamre A. G.; Jana S.; Reppert N. K.; Payne C. M.; Sørlie M. Processivity, substrate positioning, and binding: The role of polar residues in a Family 18 Glycoside Hydrolase. Biochemistry 2015, 54, 7292–7306. 10.1021/acs.biochem.5b00830. [DOI] [PubMed] [Google Scholar]
- Zees A. C.; Pyrpassopoulos S.; Vorgias C. E. Insights into the role of the (α+β) insertion in the TIM-barrel catalytic domain, regarding the stability and the enzymatic activity of Chitinase A from Serratia marcescens. Biochim. Biophys. Acta, Proteins Proteomics 2009, 1794, 23–32. 10.1016/j.bbapap.2008.09.018. [DOI] [PubMed] [Google Scholar]
- Wilson D. B. Processive and nonprocessive cellulases for biofuel production-lessons from bacterial genomes and structural analysis. Appl. Microbiol. Biotechnol. 2012, 93, 497–502. 10.1007/s00253-011-3701-9. [DOI] [PubMed] [Google Scholar]
- Heggset E. B.; Hoell I. A.; Kristoffersen M.; Eijsink V. G. H.; Vårum K. M. Degradation of chitosans with Chitinase G from Streptomyces coelicolor A3(2): Production of chito-oligosaccharides and insight into subsite specificities. Biomacromolecules 2009, 10, 892–899. 10.1021/bm801418p. [DOI] [PubMed] [Google Scholar]
- Jung W.-J.; Park R.-D. Bioproduction of chitooligosaccharides: Present and perspectives. Mar. Drugs 2014, 12, 5328–5356. 10.3390/md12115328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horn S. J.; Sørlie M.; Vaaje-Kolstad G.; Norberg A. L.; Synstad B.; Vårum K. M.; Eijsink V. G. H. Comparative studies of chitinases A, B and C from Serratia marcescens. Biocatal. Biotransform. 2006, 24, 39–53. 10.1080/10242420500518482. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.