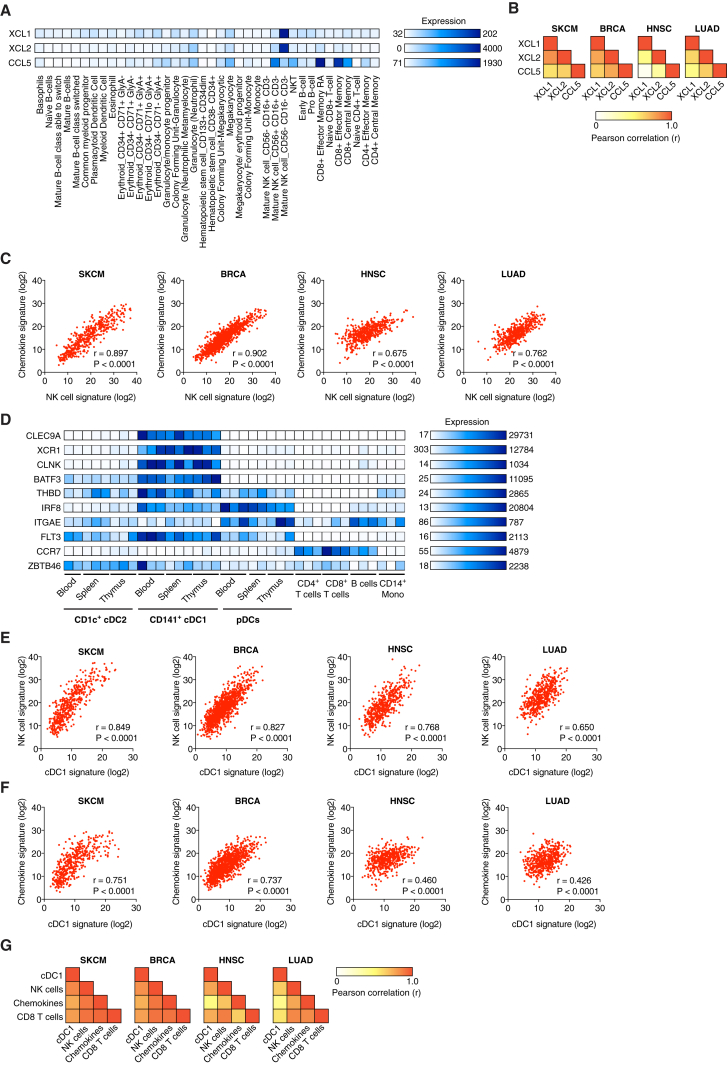
Figure 6.
Cross Correlation of Gene Signatures for NK Cells, cDC1, CCL5, XCL1, and XCL2 in Human Cancer
(A) Analysis of CCL5, XCL1, and XCL2 expression in 38 human hematopoietic cell populations based on global gene expression data (dataset from GSE24759).
(B) Heatmap showing Pearson correlation values calculated pairwise between XCL1, XCL2, and CCL5 transcript levels in human TCGA datasets for skin cutaneous melanoma (SKCM, n = 460), breast invasive carcinoma (BRCA, n = 1092), head and neck squamous cell carcinoma (HNSC, n = 518), and lung adenocarcinoma (LUAD, n = 506).
(C) Correlation between signatures for chemokines and NK cells within TCGA datasets.
(D) Identification of cDC1-specific genes in human DC subsets based on global gene expression data (dataset from GSE77671).
(E) Correlation of gene signatures specific for cDC1 and NK cells in TCGA datasets.
(F) Correlation of gene signatures for chemokines and cDC1 in TCGA datasets.
(G) Heatmap showing the Pearson correlation coefficient for the indicated gene signatures in TCGA datasets. r, Pearson correlation coefficient (r); p, p value.
See also Figure S6.