Figure S1.
Assessment of Migration Parameters in Cortical Interneurons Lacking Ccp1 Expression, Related to Figure 1
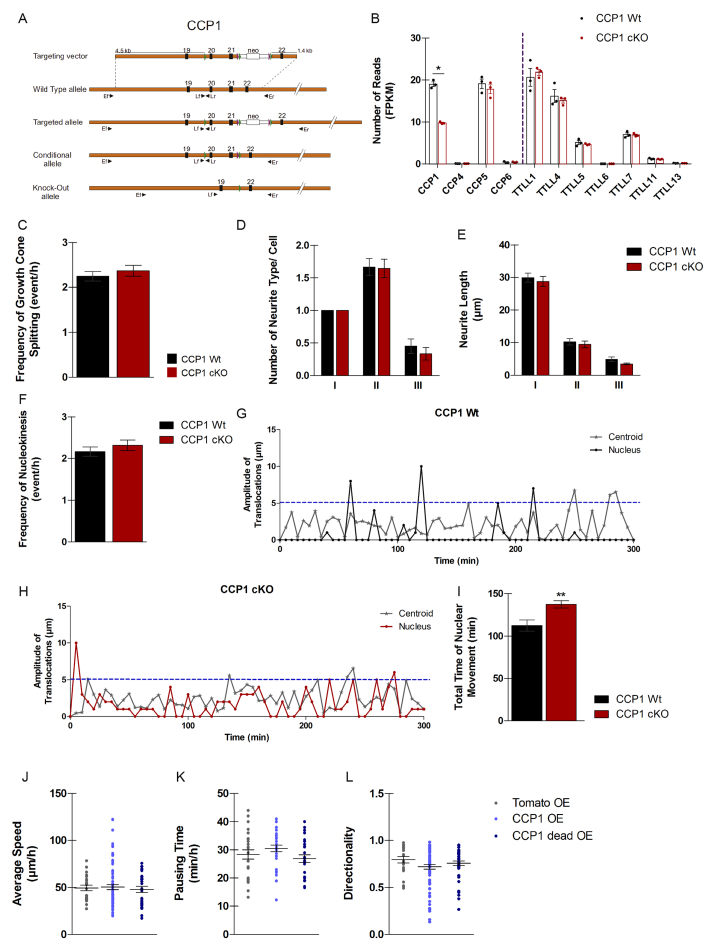
(A) Schematic representation of the targeting vector used and all possible alleles for Ccp1 gene. Orange bar: genomic DNA. Black boxes: exons with their corresponding number. Green and purple arrowheads: LoxP and Flp sequences, respectively. White bar: neo cassette, with the neomycin resistance gene (white box). Blue lines: zone of sequence homology for homologous recombination, with the corresponding size in kbp. Black arrowheads: primers used for the PCR genotyping.
(B) Normalized expression levels of CCP and TTLL mRNAs from extracted RNA of FACS E13.5 CCP1 WT and CCP1 cKO GEs. Values are expressed as FPKM, n = 3 embryos from independent female donors.
(C–F) Time-lapse experiments of CCP1 WT and CCP1 cKO cINs from explant co-cultures. Quantification of growth cone splitting (emergence of a secondary branch from the growth cone) (C), average number of different neurite types on cINs during migration (I = primary neurites, II = secondary and III = tertiary neurites) (D), neuritic length (E) and frequency of nucleokinesis (F), respectively, n = 26-52 cells from at least 3 independent cultures, two-way ANOVA.
(G and H) Representative examples of CCP1 WT and CCP1 cKO nucleus (G) and centroid displacements (H) measured by time-lapse acquisition in E13.5 GEs explants. The traveled distance between two time points is plotted and every displacement above 5μm (blue dotted line) is considered as a nucleokinesis.
(I) Quantification of the total time of nuclear movement in CCP1 WT and CCP1 cKO measured in time-lapse acquisition of E13.5 organotypic slice culture.
(J–L) Time-lapse analysis of cINs in explant co-culture showing average speed (J), pausing time (K), and directionality (L) after overexpression of Ccp1 (CCP1 OE), its catalytic dead form (CCP1dead OE), or a control (Tomato OE), n = 22-78 cells from at least 3 independent cultures, one-way ANOVA.
All graphs contain bars representing SEM.