Figure 2.
Mutations in Genes Encoding Chloroplast-Localized Proteins Cause Increased HSP70 Expression and Increased Thermotolerance
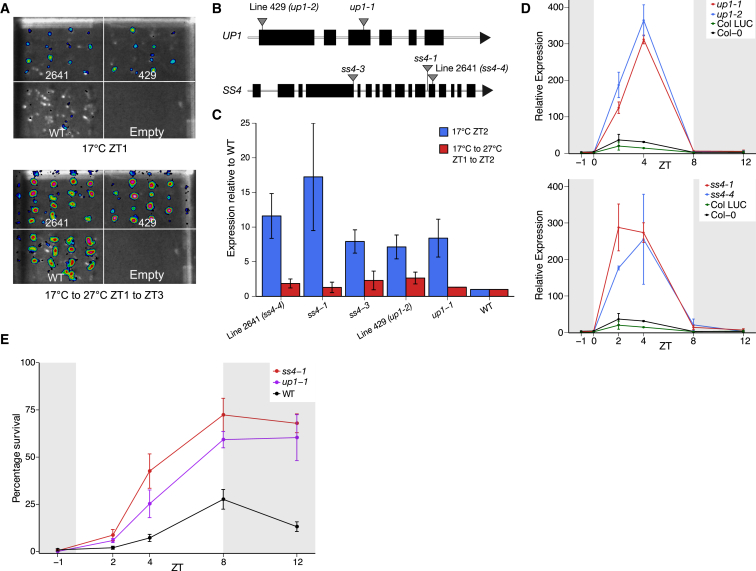
(A) False color image of LUCIFERASE activity of seedlings imaged at 17°C ZT1 and after a shift from 17°C to 27°C from ZT1 to ZT3. Two mutant lines, 2641 and 429, and WT (col-0 containing pHSP70::LUC), are shown from the same plate.
(B) Schematics of UP1 and SS4 with the position of mutations shown. Line 429 (up1-2) has a C to T substitution on chromosome (chr) V at locus 2763941 mutating Gln10 to a stop codon. Line 2641 (ss4-4) has a C to T substitution on chr IV at locus 10086122 mutating Gln872 to a stop codon.
(C and D) HSP70 expression assayed by qRT-PCR.
(C) Mutants at 17°C ZT2 and after a shift from 17°C to 27°C from ZT1 to ZT2. Expression shown relative to WT sampled from the same plate. Error bars are ± the range of two measurements (n = 2) or SEM (n = 3 to 4).
(D) up1 and ss4 mutants over a time course at 17°C in short days sampled at ZT−1, ZT0, ZT2, ZT4, ZT8, and ZT12. Error bars are ± the range of two measurements (n = 2) or SEM (n = 3).
(E) Thermotolerance of up1-1, ss4-1, and WT with plants grown at 17°C and treated at ZT−1, ZT2, ZT4, ZT8, and ZT12. Error bars are ±SEM (n = 3).