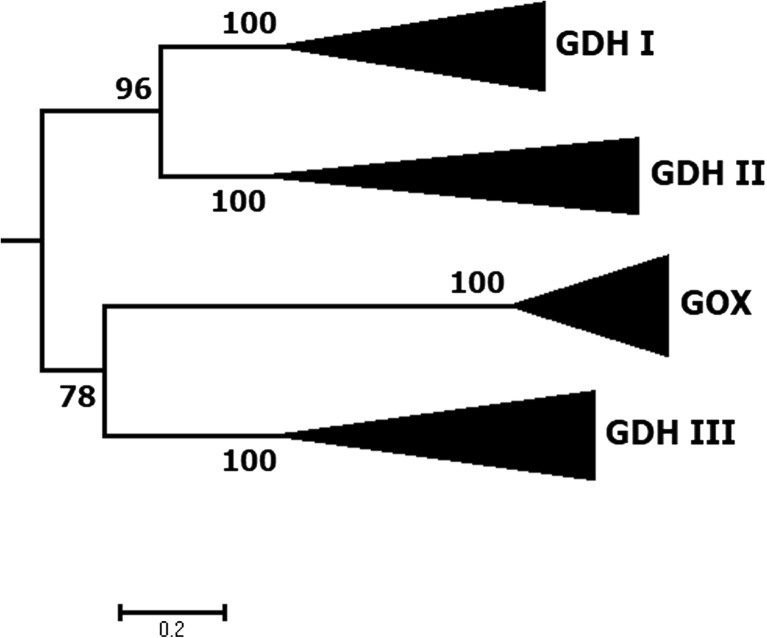
Fig. 4.
Detailed phylogeny of glucose oxidases and glucose dehydrogenases showing the newly proposed classification. An extended BLAST search resulted in numerous putative glucose oxidoreductases. The tree shown here was calculated using 28 GOx and GDH sequences representative for the clades GOx (8 sequences, 8 characterized), GDH class-I (7 sequences, 5 characterized), GDH class-II (6 sequences, 0 characterized), and GDH class-III (7 sequences, 1 characterized). The tree was rooted within AA3 family enzymes (not shown). Sequences were aligned using M-coffee (Wallace et al. 2006) with default settings. Phylogeny was inferred using PhyML (Guindon et al. 2010) and the Whelan and Goldman (WAG) amino acid substitution model (Whelan and Goldman 2001). Branch support was calculated by 100 bootstrap repetitions (values displayed in percent). The tree was visualized in MEGA 7 (Kumar et al. 2016)