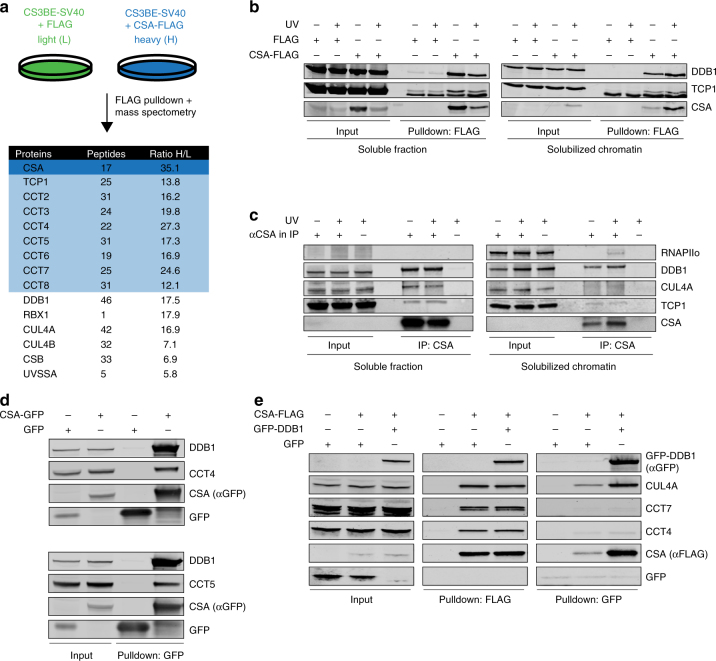
Fig. 1.
CSA interacts with chaperonin TRiC. a A SILAC-mass spectrometry approach identified all TRiC subunits as CSA-interacting proteins. CSA-deficient CS3BE-SV40 cells expressing FLAG or CSA-FLAG were cultured in medium containing light or heavy lysine and arginine isotopes, respectively. FLAG- and CSA-FLAG-interacting proteins were pulled down and samples were processed and analyzed by mass spectrometry. The table shows the number of unique peptides found for the top ranked interactors, as well as the ratio of the interactor in the CSA-FLAG pulldown to that in the control FLAG pulldown (ratio H/L). b FLAG pulldowns confirm the UV-independent interaction between CSA-FLAG and TCP1. CS3BE-SV40 cells expressing FLAG or CSA-FLAG were mock-treated or UV-C irradiated (20 J/m2). After 1 h of recovery cells were lysed and fractionated into soluble or solubilized chromatin. FLAG pulldowns using both fractions were followed by western blot analysis for the indicated proteins. c CSA co-immunoprecipitation confirms the interaction between endogenous CSA and TCP1. As in b, except that VH10-hTert cells were used and that endogenous CSA was immunoprecipitated. d GFP pulldowns confirm the interaction between CSA and TRiC subunits CCT4 and CCT5. GFP or CSA-GFP was pulled down from CS3BE-SV40 cells. e Tandem FLAG and GFP pulldowns show preferential binding of TRiC to DDB1/CUL4A/RBX1-free CSA. CSA-FLAG, GFP, and GFP-DDB1 were expressed in U2OS cells as indicated. Enrichment of CSA-interacting proteins by means of FLAG pulldowns confirmed interactions between CSA and DDB1 and CUL4A, as well as the TRiC subunits CCT4 and CCT7. Subsequently, eluted protein complexes were subjected to pulldown of GFP-DDB1, revealing an interaction with CUL4A, but not CCT4 and CCT7. Full-size scans of western blots are provided in Supplementary Fig. 7