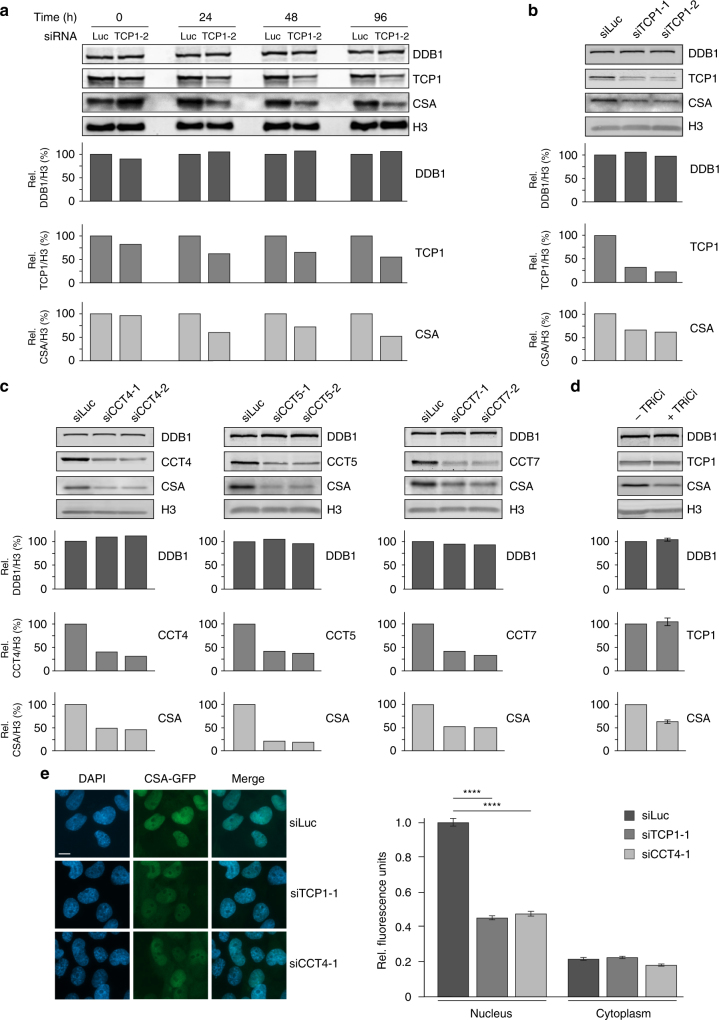
Fig. 3.
Loss of TRiC components reduces CSA stability. a Depletion of TCP1 decreases CSA protein abundance. VH10-hTert cells were transfected with the indicated siRNAs and total cell extracts were prepared at the indicated time points after siRNA transfection. Protein levels were determined by western blot analysis of the indicated proteins. H3 is a loading control. Graphs represent the ratio of protein signal intensities over H3 control signal intensities for siTCP1-treated cells relative to that for siLuc-treated control cells, which was set to 100%, at each time point. A repeat of the experiment is shown in Supplementary Figure 2a. b Depletion of TCP1 decreases CSA protein abundance. As in a, except that two different siRNAs against TCP1 were used and that protein levels were determined 72 h after siRNA transfection. A repeat of the experiment is shown in Supplementary Figure 2b. c Depletion of CCT4, CCT5, or CCT7 decreases CSA protein abundance. As in a, except that CCT4, CCT5, or CCT7 siRNAs were used and that protein levels were determined 72 h after siRNA transfection. A repeat of the experiment is shown in Supplementary Figure 2c. d TRiC inhibition decreases CSA protein abundance. VH10-hTert cells were treated with DMSO or an inhibitor against the TRiC subunit TCP1 (TRiCi). Protein levels were determined after 72 h of treatment. e TCP1 or CCT4 loss decreases CSA-GFP protein abundance in the nucleus. TCP1 or CCT4 was depleted from CSA-GFP expressing CS3BE-SV40 cells using the indicated siRNAs. Nuclear and cytoplasmic CSA-GFP levels were analyzed and quantified by fluorescence microscopy and ImageJ. GFP signal intensities were normalized to the average nuclear signal in siLuc-treated cells. Data represent mean ± SEM of 190 cells quantified in two independent experiments. p-Values were derived from an unpaired t-test. Length of scale bar: 10 µm. Full-size scans of western blots are provided in Supplementary Fig. 8