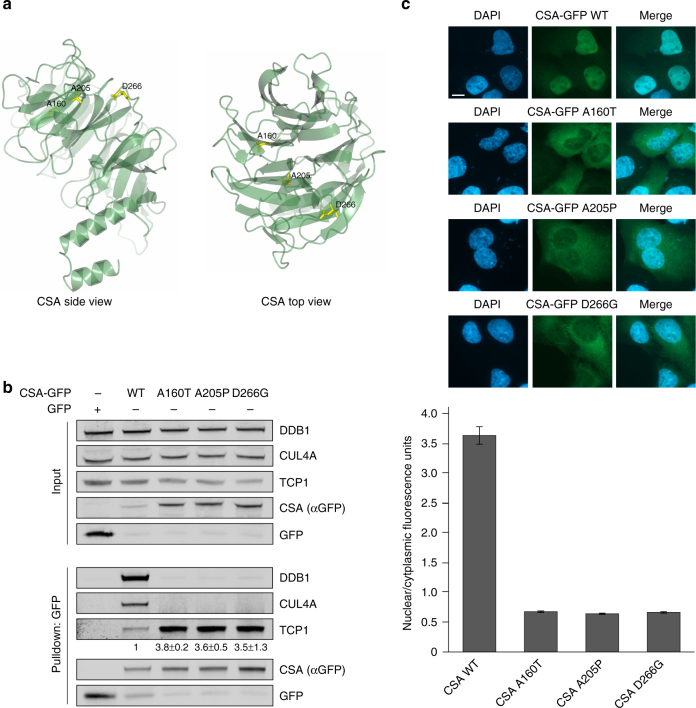
Fig. 7.
Patient mutations in CSA cause increased TRiC binding. a Side and top view of CSA. Residues Ala160, Ala205, and Asp266 that have been found mutated in Cockayne syndrome patients are shown in yellow. Visualization was done in ccp4mg using structure 4a11 from the PDB. b CSA harboring patient mutation A160T, A205P, or D266G shows increased binding to TRiC and failure to be incorporated into the CRLCSA complex. CSA-GFP WT and CSA-GFP containing the indicated mutations were pulled down from U2OS cells. Protein levels were determined by western blot analysis. The signal intensity ratio of TCP1 over the CSA-GFP mutant relative to that of TCP1 over CSA-GFP WT, which was set to 1, is shown as the mean ± SEM of two independent experiments. c CSA A160T, A205P, and D266G show predominant cytoplasmic localization. CSA-GFP WT and CSA-GFP containing the indicated mutations were expressed in U2OS. Mean nuclear and cytoplasmic GFP intensities were analyzed and quantified by fluorescence microscopy and ImageJ. For each cell, the nuclear/cytoplasmic ratio was calculated. Data represent mean ± SEM of 100 cells quantified in two independent experiments. Length of scale bar: 10 µm. Full-size scans of western blots are provided in Supplementary Fig. 10