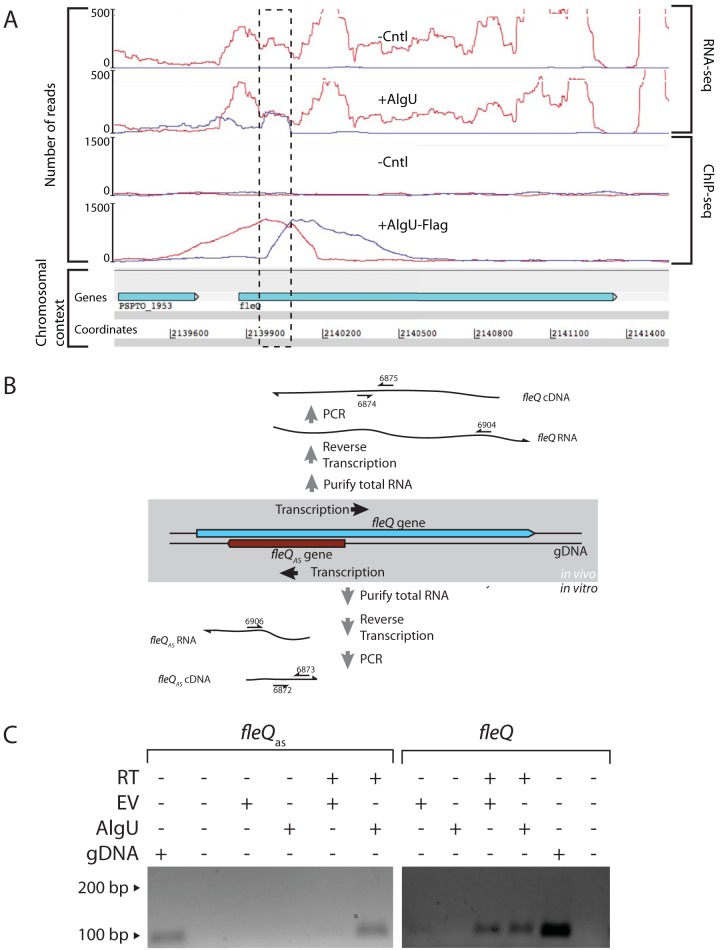
FIG 1.
Analysis of AlgU-dependent antisense transcript expressed within the fleQ gene. (A) RNA-seq and ChIP-seq results at the fleQ locus. Strand-specific RNA-seq analysis of transcriptomes produced by P. syringae pv. tomato DC3000 ΔalgU mucAB cells containing an AlgU expression construct (+AlgU) or the empty-vector control (-Cntl) show the location of antisense transcription in the fleQ gene (dashed box). ChIP-seq analysis of P. syringae pv. tomato DC3000 ΔalgU mucAB cells containing an AlgU-Flag expression construct (+AlgU-Flag) or the empty-vector control (-Cntl) shows AlgU-Flag-dependent enrichment of sequences directly upstream of the location of fleQas transcription. Experimental details and complete data sets for the AlgU RNA-seq and ChIP-seq results are available in reference 22. The number of reads on the y axis indicates the number of sequence reads mapped per nucleotide position. The red and blue profiles indicate sequences matching the sense and antisense strands, respectively. (B) Strand-specific RT-PCR was used to test for the fleQas transcript in cells containing either the AlgU expression plasmid or the empty-vector control. PCR amplification required the product of strand-specific reverse transcription of the fleQas or the fleQ transcript. Primers complementary to either the fleQas or the fleQ transcript were used to reverse transcribe a segment of each transcript that served as the template in a PCR to detect the cDNA products. The numbered arrows refer to oligonucleotide primers (see Table S3 in the supplemental material). (C) Agarose gel showing reverse transcriptase (RT)-dependent PCR products of fleQas, which were obtained only from cells expressing AlgU. In contrast, the fleQ sense transcripts were detected in cells containing AlgU expression and the empty vector (EV). P. syringae pv. tomato DC3000 genomic DNA (gDNA) was a positive control for the gene-specific PCR.