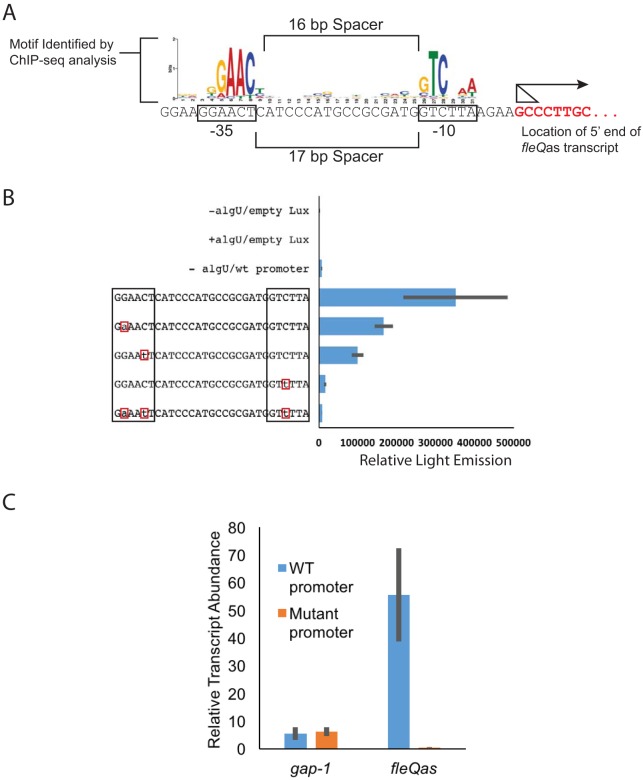
FIG 3.
An AlgU-dependent promoter controls fleQas expression. (A) Comparison of the nucleotide sequence directly upstream of the predicted fleQas transcription start site to that of the AlgU-dependent promoter reported in reference 22. The arrow shows the direction of transcription, and the triangle shows the range of potential start sites. (B) Luciferase reporter expression of wild-type (wt) and mutant fleQas promoter::lux fusions in the pBS58 vector. Empty Lux represents the reporter activity of the empty lux reporter construct in the presence and absence of an AlgU expression vector. The lux reporter vector containing the wild-type fleQas promoter is indicated by “wt promoter” or by the unchanged full promoter sequence. −algU/wt promoter, Lux expression from the wild-type promoter in the absence of AlgU. The lux reporter vectors containing the unchanged or changed promoter sequences were expressed in the presence of AlgU (indicated by the promoter sequences that show altered nucleotides [boxed in red]). Relative Light Emission indicates the light emission/OD600 unit. The sequences of the wild-type promoter and altered nucleotides are indicated. (C) qRT-PCR analysis of fleQas expression in AlgU-expressing strains (P. syringae pv. tomato DC3000 ΔalgU mucAB plus pEM53) versus that with the empty-vector control (P. syringae pv. tomato DC3000 ΔalgU mucAB plus pJN105) with the wild-type or mutant fleQas promoter. Averages and standard deviations from three biological replicates are shown for Lux and qRT-PCR expression analyses.