Abstract
yonT/SR6 is the second type I toxin-antitoxin (TA) system encoded on prophage SPβ in the B. subtilis chromosome. The yonT ORF specifying a 58 aa toxin is transcribed on a polycistronic mRNA under control of the yonT promoter. The antitoxin SR6 is a 100 nt antisense RNA that overlaps yonT at its 3′ end and the downstream gene yoyJ encoding a second, much weaker, toxin at its 5′ end. SR6 displays a half-life of >60 min, whereas yonT mRNA is less stable with a half-life of ≈8 min. SR6 is in significant excess over yonT mRNA except in minimal medium with glucose. It interacts with the 3′ UTR of yonT mRNA, thereby promoting its degradation by RNase III. By contrast, SR6 does not affect the amount or half-life of yoyJ mRNA. However, in its absence, a yoyJ overexpression plasmid could not be established in Bacillus subtilis suggesting that SR6 inhibits yoyJ translation by directly binding to its ribosome-binding site. While the amounts of both yonT RNA and SR6 were affected by vancomycin, manganese, heat-shock and ethanol stress as well as iron limitation, oxygen stress decreased only the amount of SR6.
Keywords: yonT/SR6, sRNA, small regulatory RNA, Bacillus subtilis, type I toxin-antitoxin system, multistress responsive TA system
1. Introduction
To date, six types of toxin–antitoxin systems are known (reviewed in [1,2]). All these systems comprise two genes, one encoding a stable toxin, the other an unstable antitoxin that neutralizes the toxin action. Whereas in type I and III TA systems, the antitoxin is a small RNA, in type II, IV, V and VI systems toxin and antitoxin are proteins. In the recently described type VI system, the toxin blocks replication elongation by directly binding to the β sliding clamp DnaN, whereas the antitoxin promotes toxin degradation by ClpXP [3]. Type I antitoxins interact with their toxin mRNAs that mostly code for small hydrophobic peptides recruited to the cell membrane, but also for RNA or DNA cleaving enzymes (rev. in [4]). The first TA systems have been found on plasmids, where they act as post-segregational killing (PSK) systems (rev. in [5]). Subsequently, a multitude of TA systems were discovered on bacterial chromosomes (e.g., [6]).
Type I TA systems have been found in many Gram-positive and Gram-negative species, and some of them were investigated in great detail (e.g., hok/Sok, rev. in [5]; or fst/RNAI, rev. in [7,8]). TA loci are arranged as overlapping, convergently transcribed gene pairs or as divergently transcribed pairs located apart (rev. in [4]). In the first ones, the antitoxin is a cis-encoded antisense RNA (e.g., hok/Sok, rev. in [9]; txpA/RatA [10,11]; aapA1/IsoA1 [12]), in the latter ones, it is a trans-encoded sRNA (e.g., tisB/IstR1, rev. [13]; zor/Orz, ibs/Sib and shoB/OhsC, rev. in [14]). In some cases, deletion of the antitoxin gene results in cell lysis [15], in others, only toxin overexpression impairs growth (zor/Orz, tisB/IstR1). Whereas TA pairs on plasmids act as PSK required for plasmid maintenance (e.g., hok/Sok on E. coli plasmid R1 or fst/RNAII on E. faecalis plasmid pAD1, rev. in [7,8]), the biological function of many chromosomal TA loci is still poorly understood. E. coli symE/SymR was proposed to be involved in recycling of damaged RNA generated upon SOS stress (rev. in [16]), dinQ/AgrB in chromosome stability [17] and ralR/RalA in resistance against cell-wall inhibiting antibiotics [18]. By contrast, for E. coli tisB/IstRI and hokB/SokB as well as Streptococcus mutans fstSm/SrSm, a role in persister formation has been demonstrated (rev. in [4]), [19]. B. subtilis txpA/RatA probably helps—similar to plasmid-encoded PSK systems—to maintain the skin element, which is excised from the chromosome during sporulation, thereby allowing reconstitution of a functional sigK gene [10]. In other cases, it was speculated that toxin expression might be advantageous upon stress, as shown for some type II TA systems (rev. in [9]).
In B. subtilis, 14 type I TA systems were proposed [20], and five of them are located on prophage regions. Three of them—txpA/RatA [10,11], bsrG/SR4 [15] and bsrE/SR5 [21,22]—have been investigated in more detail. bsrG/SR4 located on SPβ prophage is the first discovered temperature-sensitive type I TA system, as bsrG RNA is rapidly degraded at 48 °C. SR4 is a bifunctional antitoxin that promotes bsrG RNA degradation by recruiting RNase III, and additionally induces a structural change around the bsrG RBS that further obstructs translation [23]. The toxin BsrG (38 aa) is recruited to the cell membrane, but does not dissipate the membrane potential. Instead, it generates membrane invaginations causing delocalization of the cell-wall synthesis machinery, which finally results in autolysis [24]. bsrE/SR5 is a multistress-responsive TA system encoded on the prophage-like region P6 [21]. The secondary structures of bsrE RNA and bsrG RNA are very similar, and binding between toxin mRNA and the cognate antitoxin occurs in both systems via three subsequent steps [22]. However, SR5 is—in contrast to SR4—a monofunctional antitoxin that only promotes degradation of its bsrE toxin mRNA by recruiting RNase III. Both bsrE mRNA and SR5 are rapidly degraded by RNase Y upon ethanol stress. Whereas bsrE RNA is influenced by temperature shock and alkaline stress, SR5 is rapidly degraded under O2 limitation and affected by pH stress and iron limitation [21].
Here, we confirm experimentally previous data [6,20] that yonT/SR6 is the second type I TA system located on the SPβ prophage of the B. subtilis chromosome. We identified the transcription start points of yonT mRNA and SR6 and analyzed the expression profiles of both RNAs in different media and under stress conditions. Furthermore, we determined the strength of the yonT and sr6 promoters. In addition, we calculated the half-lives of toxin mRNA and RNA antitoxin. YonT proved to be so toxic that only a mutated yonT gene containing a stop instead of a start codon could be cloned in E. coli and established in B. subtilis. Determination of the intracellular concentrations of both complementary RNAs showed that SR6 is present at least at 10- to 25-fold higher concentrations than yonT or yoyJ mRNA under most growth conditions. We demonstrate by Northern blotting that SR6 acts by promoting yonT mRNA degradation. Furthermore, we provide indirect evidence that yoyJ whose RBS is complementary to the 5′ end of SR6 is a second, albeit weaker, toxin that might be also inhibited by SR6. A BLAST search for homologues of yonT, yoyJ and sr6 revealed that yonT and sr6 are encoded in five Bacillus species, whereas the yoyJ gene or its promoter was mutated, so that no functional protein can be expressed.
2. Results
2.1. yonT Is Located in an Operon, and Transcripts of Different Length Originate from pyonT
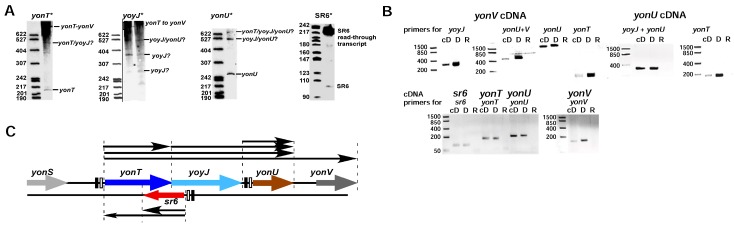
First, we performed Northern blots with total RNA isolated from B. subtilis wild-type strain DB104 grown in complex TY medium and probes against yonT RNA, yonU RNA and antisense-yonT RNA (renamed as SR6). Surprisingly, we detected yonT RNA species of different length from the predicted 217 over 527 to 1 kb and longer (Figure 1A) suggesting that yonT is located in an operon with adjacent genes. SR6 of the expected length of ≈100 nt was observed, but in addition at OD600 = 3.0 a longer SR6 transcript of ≈215 nt was detected. Second, to investigate if yonT is transcribed under an own promoter pyonT or from the upstream yonS promoter pyonS and is located in an operon together with the downstream genes yoyJ, yonU and yonV, we performed RT PCR with the same total RNA treated with DNase and primers for the detection of yonT, yonU, yonV and yoyJ transcripts. In parallel, this RNA was employed for RT-PCR with primers against SR6. Chromosomal DNA from strain DB104 served as positive control for the expected PCR fragments. Primers against gapA RNA were used as control for the purity and applicability of the isolated DNase-treated RNA (Figure S1). As shown in Figure 1A and supported by the RT-PCR results in Figure 1B, one long mRNA comprising yonT, yoyJ, yonU and yonV was detectable that is transcribed from promoter pyonT. Furthermore, a short 217 nt monocistronic yonT mRNA and a bicistronic yoyJ/yonU transcript—both apparently processing products of the long RNA—are visible. In addition, a tricistronic yonT/yoyJ/yonU transcript was found. Furthermore, a monocistronic yonU transcript was observed that might be transcribed from its own promoter downstream of yoyJ and terminated downstream of yonU. SR6 of the expected size could also be confirmed. An additional Northern blot with riboprobes against the upstream and downstream region of SR6 revealed that the ≈215 nt SR6 species observed at the transition to stationary growth phase at OD600 = 3–4 indeed originated from pSR6 and extended at least towards pyonT. No signal was obtained with a probe against a sequence upstream of SR6 (not shown). Figure 1C summarizes all transcripts detected in Northern blots including the long transcript confirmed by RT-PCR in the region between pyonT and pyonV.
Figure 1.
Determination of transcripts within the yonSTUV operon by RT-PCR. B. subtilis DB104 was grown in TY medium until OD600 = 3.0 (transition phase), total RNA isolated, treated with DNase and used as template for RT-PCR. (A) Northern blots. B. subtilis strain DB104 was grown at 37 °C in TY, aliquots taken at different times, immediately frozen in liquid nitrogen and later on used for the preparation of total RNA. RNA was separated on 6% denaturing PAA gels, blotted onto nylon membrane and hybridized with riboprobes against yonT mRNA, yoyJ mRNA, yonU mRNA and SR6. Autoradiograms of the corresponding gels are shown; (B) Original data of RT-PCR. B. subtilis DB104 was grown in TY medium until OD600 = 3.0 (transition phase), total RNA isolated, treated with DNase and used as template for RT-PCR. PCR products were analysed in ethidium-bromide stained 3% agarose gels. cD, cDNA; D, genomic DNA; R, total RNA; (C) Graphic summary of the transcripts in the yonSTUV region based on the RT-PCR data and Northern blots.
2.2. Mapping of 5′ and 3′ Ends of yonT RNA, SR6 and yonU RNA
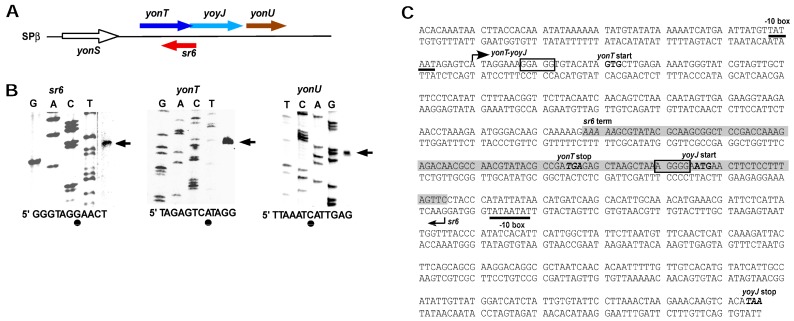
An sRNA antisense to yonT was first detected by Fozo et al. [6]. To conform to the other known B. subtilis TA systems [20], we renamed antisense-yonT as SR6 (see above) and determined the transcriptional start sites of yonT mRNA, SR6 and yonU mRNA. The latter was included to confirm that yonU is transcribed from an own promoter (−10 box: TATAAT 120 bp upstream of the TTG start codon) as depicted in Figure 1C. Figure 2A presents the chromosomal location of the yon region and Figure 2B the results of the 5′ end mapping. No typical rho-independent terminators were found downstream of yonT, yoyJ and yonU, indicating that the monocistronic yonT, yonU and yoyJ transcripts are most likely processing products. As S1 mapping also did not allow the exact determination of the 3′ ends, the RNA sizes calculated by Northern blotting (see Figure 1A and below) were used to narrow down the 3′ ends of monocistronic yonT RNA, monocistronic yonU RNA and SR6. All three RNAs are transcribed from σA promoters, and psr6 and pyonT have extended −10 boxes. SR6 and yonT overlap by ≈65 nt at their 3′ ends, whereas sr6 and yoyJ overlap by ≈30 to 40 nt at their 5′ ends. Figure 2C displays the complete nucleotide sequence of the yonT/sr6/yoyJ locus.
Figure 2.
Location of the yonT/sr6/yoyJ/yonU locus and determination of transcription start sites. (A) Schematic representation of the location of the yonT/sr6/yoyJ/yonU locus on the B. subtilis chromosome. The directions of transcription are indicated by arrows; (B) Mapping of the 5′ ends of yonT mRNA, yonU mRNA and SR6. Sanger sequencing reactions of heterologous genes were used as references to calculate the location of the transcription start signals. Outer right lanes show primer extension reactions on total RNA of B. subtilis DB104 with 5′-labelled primer SB2648 (sr6), SB2646 (yonT), and SB2647 (yonU) loaded in parallel to the sequencing reactions (transcription start sites are indicated by arrows). Below: sr6, yonT and yonU sequences around the TSS are shown. Dots symbolize the mapped TSS; (C) Sequence of the yonT/yoyJ/sr6 locus. −10 boxes of the yonT and the sr6 promoters are underlined. Start and direction of transcription are indicated by arrows and transcription termination is indicated by ‘term’. The yonT/SR6/yoyJ complementary region is highlighted in light grey. Start and stop codons of the yonT and yoyJ ORFs are in bold and the putative SD sequences are boxed.
2.3. Expression Profiles of yonT mRNA, yoyJ mRNA, yonU mRNA and SR6 in Complex TY and Minimal CSE Medium
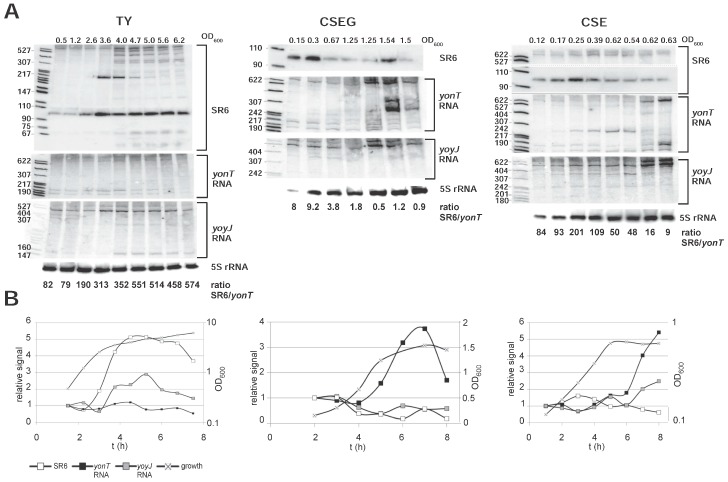
The expression patterns of yonT RNA, yoyJ RNA and SR6 were analyzed by Northern blotting after growth of B. subtilis strain DB104 at 37 °C in either complex TY or minimal CSE medium with glucose (CSEG) or without glucose (CSE). The Northern blots are displayed in Figure 3A and graphs for the calculation of RNA amounts in Figure 3B. To determine the ratio between yonT toxin mRNA and antitoxin SR6, defined amounts of in vitro synthesized yonT RNA and SR6 were loaded onto the same gel and hybridized with the same riboprobes (entire blots are shown in Figure S2). The calculations of the SR6/yonT RNA ratios over growth in all three media are summarized in Figure 3A below the autoradiograms of the Northern blots.
Figure 3.
Expression profiles and concentrations of yonT mRNA, yoyJ mRNA and SR6 in complex TY and minimal CSE medium. (A) Northern blots. B. subtilis strain DB104 was grown at 37 °C in TY or CSE medium, and RNA isolation, Northern blotting and reprobing performed as in Figure 1. For the correction of loading errors, filters were reprobed with [γ-32P] ATP-labeled oligonucleotide C767 specific for 5S rRNA. Autoradiograms of the corresponding gels are shown. The SR6/yonT RNA ratios shown below the Northern blots are derived from the calculations represented by graphs in (B). Graphic representation of the amounts of yonT RNA, yoyJ RNA and SR6 based on the quantification of the gels shown in (A) and Figure S2. The corresponding growth curves are shown using the Y axis at the right side of each graph.
In TY medium, the amount of yonT mRNA was nearly unchanged over time. By contrast, the amount of yoyJ mRNA was two- to threefold higher in stationary phase. The amount of the antitoxin SR6 increased 5-fold from early log until transition to stationary growth phase and then slightly decreased in stationary phase (Figure 3A). Here, the SR6/yonT RNA ratio increased from 82 in log phase to 313 in transition phase and was 574 in late stationary phase indicating that under these conditions, toxin expression is strongly inhibited. By contrast, in CSEG medium, the amount of yonT RNA increased about four- to five-fold until stationary phase, whereas the amount of SR6 decreased at the same time about four- to five-fold, resulting in lowest SR6/yonT RNA ratios between 0.46 and 1.22 in stationary phase. This indicates that from transition phase until late stationary phase, yonT might be expressed in some cells. The amount of yoyJ RNA was nearly constant over growth. In CSE medium, the amount of SR6 was more or less constant over growth, whereas the amount of yonT mRNA increased in stationary phase nearly six-fold. Therefore, the SR6/yonT mRNA ratio decreased from logarithmic to stationary phase from 80–100 to ≈ 9, but was always in a range that would not allow toxin expression. The amounts of yoyJ RNA increased about two-fold towards stationary phase.
In summary, we can conclude that the amount of SR6 increased only in TY medium significantly towards stationary phase, whereas it was nearly constant in CSE. As, however, the amount of yonT RNA displayed a 4 or 5.5-fold increase in CSEG and CSE, respectively, the SR6/yonT ratio decreased in these media. But only in CSEG this ratio fell to about 1, so that only in this medium toxin expression might be expected.
2.4. The sr6 Promoter Is Weaker than the yonT Promoter
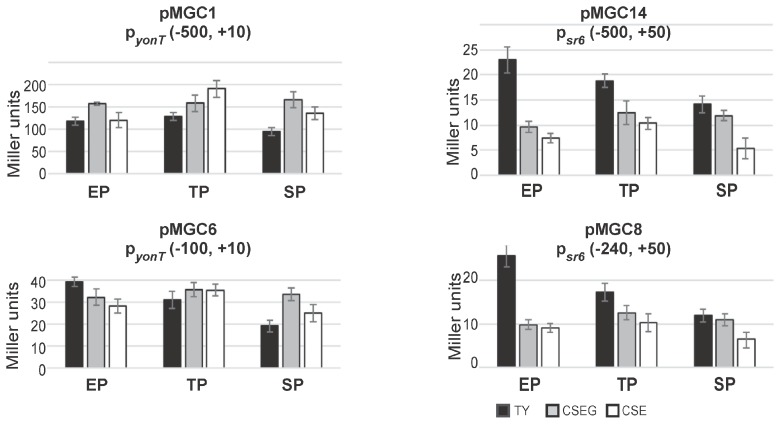
To analyze activities of the promoters pyonT and psr6, β-galactosidase measurements with transcriptional lacZ fusions were performed in TY, CSE and CSEG in early and late logarithmic and in stationary growth phase (Figure 4). To this end, transcriptional lacZ fusions comprising 500 bp upstream of the -35 boxes of pyonT (plasmid pMGC1) and psr6 (pMGC14) integrated into the amyE locus of strain DB104. Two additional plasmids, pMGC6 and pMGC8 carrying only 100 or 240 bp upstream of the -35 boxes, were also constructed.
Figure 4.
Activities of the yonT promoter and the sr6 promoter in TY (black), CSEG (grey) and CSE (white) medium over growth. B. subtilis strains generated by integration of the indicated vector fragments into the amyE locus of DB104 were grown in TY, CSEG or CSE medium. In early exponential phase (EP), late exponential phase/transition phase (TP) or stationary phase (SP) samples were withdrawn and used for β-galactosidase measurements. Averages of three independent measurements with standard deviations are shown. Vector pMG16 without a promoter insert yielded in all tested media values between 1 and 3 Miller units.
As shown in Figure 4, between media and growth phases, no significant differences in the activities of pyonT could be found. The pyonT-lacZ fusion with the 500 bp promoter upstream region revealed in all three growth phases activities between 100 and 200 MU. In TY and CSEG medium, only a slight alteration during growth was observed, whereas in CSE medium a two-fold increase only in late exponential phase was determined. By contrast, the pyonT-lacZ fusion with the short (100 bp) promoter upstream region (pMGC6) revealed with 20–40 MU four- to five-fold lower activities. In TY, activities decreased from exponential to stationary phase about 2-fold, whereas in CSEG and CSE no significant alterations were observed. The activities of psr6 were with 15 to 25 MU very low. This was unexpected, as type I antitoxins are usually transcribed from rather strong promoters (e.g., a psr4-lacZ fusion yielded between 750 and 3000 MU [15]). However, measurements with an empty vector yielding 0.5 MU (early log phase) to 3 MU (stationary phase) confirmed that psr6 was not inactive. The constructs pMGC14 (−500, +50) and pMGC8 (−240, +50) displayed in TY a two-fold activity decrease from early log to stationary phase, whereas in CSE and CSEG only marginal changes were observed. Similar low activities (15–27 MU) were measured for pyonU in all growth phases and media (not shown).
In summary, the measured activities of psr6 were in all growth media and stages between 20- and two-fold lower than those of pyonT. The activities of pyonT were four- to five-fold higher in constructs with a 500 bp promoter upstream region compared to those with only a 100 bp promoter upstream region suggesting a possible regulation by a transcriptional activator. Whereas pyonT activities were essentially independent of the growth medium, psr6 activities were—at least in early log phase—highest in TY medium.
2.5. Overexpression of yoyJ Impairs the Establishment of Transformants in B. subtilis Indicating That It Is Also a Toxin
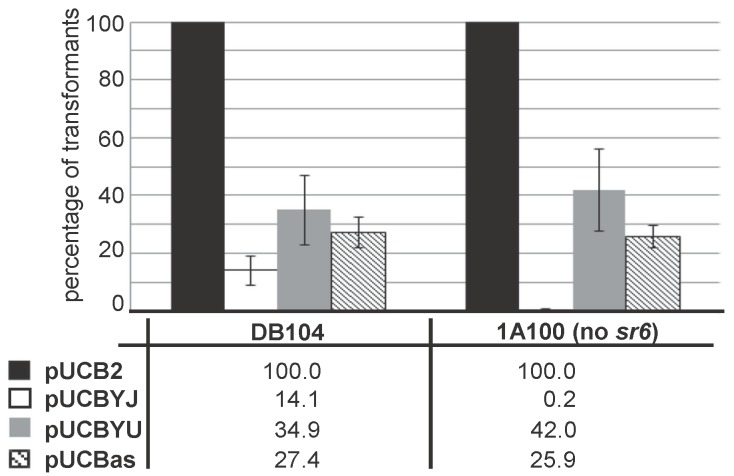
As SR6 is not only complementary to the 3′ region of yonT mRNA, but also to the 5′ region including the RBS of yoyJ (see Figure 2A,C), it was previously hypothesized [20] that YoyJ might be also a toxin, and its expression inhibited by SR6. To investigate this issue, we constructed pUCBYJ, a multicopy E. coli/B. subtilis shuttle vector (≈50 copies/cell) that allows for overexpression of yoyJ in both E. coli and B. subtilis. Already upon transformation of E. coli we observed rather small transformant colonies that grew much slower than those of pUCBas used for overexpression of the complete sr6 gene. Therefore, we decided to compare the transformation efficiency of pUCBYJ in B. subtilis with that of pUCBas (encoding sr6) and pUCBYU encoding yonU that is not proposed to be a toxin. As control served the empty vector pUCB2. In parallel we used B. subtilis strain 1A100 lacking the prophage SPβ that does not encode yonT/yoyJ/sr6 and DB104 containing SPβ, and, thus, expressing the entire yonSTUV operon including sr6. As shown in Figure 5, the transformation efficiencies of pUCBas and pUCBYU were comparable in both strains, while the empty vector pUCB2 yielded about 5 times more colonies. By contrast, pUCBYJ could not be established in strain 1A100 that does not express the antitoxin SR6, whereas it yielded a few transformants in DB104. When we grew three DB104(pUCBYJ) transformants alongside three DB104(pUCBas) transformants, they did not display altered growth (not shown). Initially we suspected that they might have undergone mutations in the yoyJ gene, but sequencing of 10 clones demonstrated that this was not the case.
Figure 5.
Establishment of transformants overexpressing yoyJ in B. subtilis in the presence and absence of sr6. B. subtilis strains DB104 (wild-type) and 1A100 (ΔSPβ, i.e., Δ[yonT/yonY/sr6]) were transformed with 5 μg purified plasmids pUCB2 (empty vector control), pUCBYJ (overexpression of yoyJ), pUCBYU (overexpression of yonU) and pUCBas (overexpression of sr6), transformants selected for kanamycin resistance and counted. The number of pUCB2 transformants was set to 100%. Shown data are the average of four independent experiments.
In summary, we can conclude that YoyJ is, in addition to YonT, most probably also a toxin, although its toxicity is much lower than that of YonT.
2.6. SR6 Is Significantly More Stable than yonT mRNA
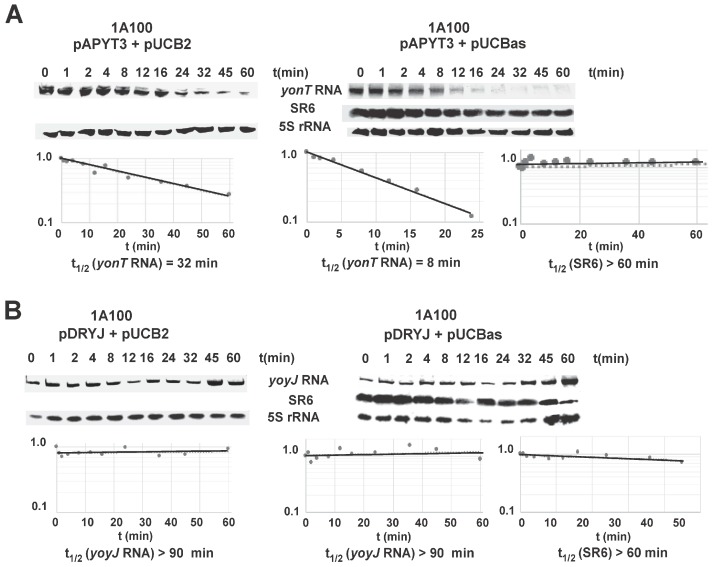
Half-lives of yonT RNA and SR6 were determined in B. subtilis 1A100 in TY at OD600 = 3.0, i.e., at transition to stationary phase when both long and short SR6 species were visible. A yonT overexpression plasmid was used, as the signals from the chromosomal yonT gene in Northern blots were extremely weak (see Figure 3) impeding an exact calculation of a progressive decrease over time. The yonT RNA proved to be unstable with a half-life of 8 min (Figure 6A). The half-life of the 100 nt SR6 species was ≈60 min, whereas the 215 nt SR6 species was extremely stable with a half-life of >120 min.
Figure 6.
Determination of the half-lives of yonT mRNA and SR6 (100 nt) in complex TY medium. Half-lives were determined as described (Materials and Methods). Samples were taken at the indicated times after addition of 200 μg/mL rifampicin at OD600 = 3.5. [α-32P]-UTP-labeled riboprobes for yonT RNA and SR6 were used. Reprobing was performed as in Figure 3. Autoradiograms of the Northern blots are shown. Indicated half-lives are averaged from three independent determinations. (A) Determination of half-lives of yonT RNA expressed from overexpression plasmid pAPYT3; (B) Determination of half-lives of yoyJ RNA expressed from overexpression plasmid pDRYJ1. Strain 1A100 (no sr6 gene) containing either pAPYT3 or pDRYJ1 combined with either pUCB2 (vector pUCB2) or pUCBas (pUCB2 with sr6 gene under own promoter) were grown in TY with IPTG to induce yonT and yoyJ transcription, respectively.
2.7. SR6 Promotes Degradation of yonT RNA, but Does Not Affect the Stability of yoyJ RNA
In many type I TA systems, the antitoxin promotes degradation of the toxin mRNA. To determine whether SR6 affects stability of yonT mRNA, SR6 was expressed in strain 1A100 from multicopy plasmid pUCBas and the empty vector pUCB2 served as negative control. The toxin gene yonT was expressed in the same strain from the chromosome using an integration vector (pAPYT3). Since we could not clone wild-type yonT in any tightly regulated inducible, high or low copy vector in E. coli (which is the prerequisite for analyzing yonT expression in B. subtilis 1A100)—even in the presence of a plasmid expressing sr6—we had to resort to cloning of a yonT gene with a start- to stop codon mutation (pAPYT3). In the absence of SR6, the half-life of yonT mRNA was ≈32 min, and it decreased to ≈8 min in the presence of SR6 (Figure 6A). With a half-life of >60 min, the stability of SR6 was not affected even by overexpression of yonT mRNA. This was not unexpected as we determined a high (300–350-fold in transition phase) excess of SR6 over yonT RNA during growth in TY medium (Figure 3A). As SR6 might also affect the half-life of yoyJ mRNA encoding the second toxin (see above), wild-type yoyJ was expressed from an integration vector (pDRYJ) in strain 1A100 either in the presence (pUCBas) or absence (pUCB2) of SR6. In both cases, yoyJ RNA was very stable with a half-life of >90 min, whereas the half-life of SR6 was not affected by yoyJ mRNA (Figure 6B).
Therefore, we can conclude that SR6 promotes degradation of yonT mRNA whereas it does not affect the stability of yoyJ mRNA.
2.8. The Amounts of yonT RNA and SR6 Are Affected by Different Stress Conditions
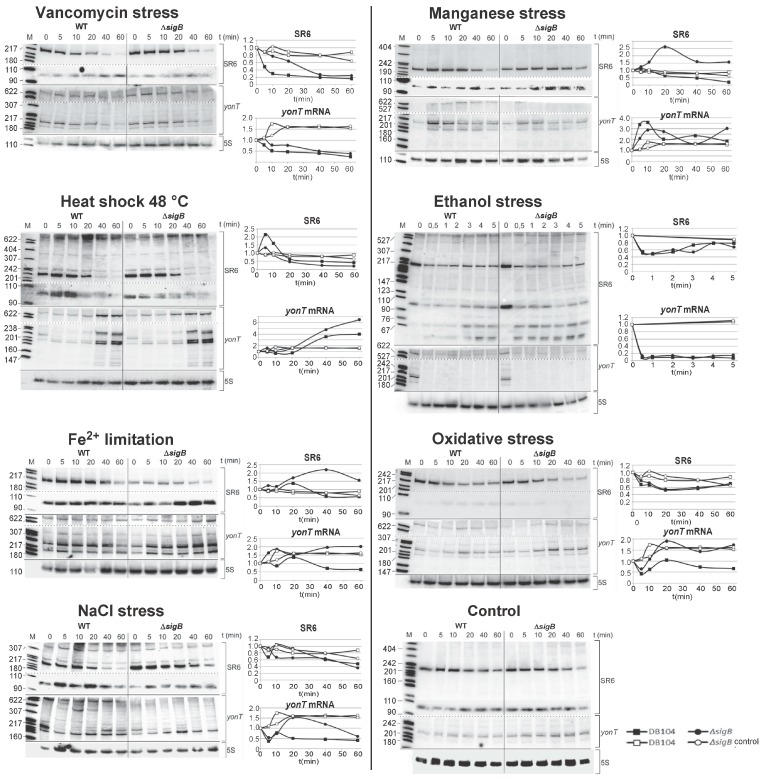
To identify factors that might impact yonT or sr6 expression, DB104 was cultivated until onset of stationary phase in TY medium, different stress conditions were applied, time samples taken over 60 min, total RNA prepared and subjected to Northern blotting (Figure 7). To find out if the stress effects depend on the alternative sigma factor σB, a sigB knockout strain was used in parallel. The following stress conditions, which did not impair cell viability, were tested as described in Materials and Methods: Cell wall stress (vancomycin), manganese stress, heat-shock, ethanol stress, iron limitation, oxidative and salt stress as well as anaerobic stress with nitrate respiration. The latter stress had no pronounced effects on either RNA (not shown). The effects of the other stress conditions are discussed below and summarized in Figure 7.
Figure 7.
Stress conditions that influence the amounts of yonT RNA and SR6. B. subtilis strain DB104 was cultivated in TY at 37 °C until onset of stationary phase, stress factors applied as described in Materials and Methods, time samples withdrawn after indicated times, RNA prepared and subjected to Northern blotting as in Figure 1. Wild-type and ΔsigB strains were compared. Analysis of cell wall (vancomycin) stress, heat shock, Fe2+ limitation, salt, manganese, ethanol stress, and oxygen stress. Furthermore, a control blot without applied stress factors at 37 °C is included. Next to each Northern blot, the graphic representation of the stress effects on yonT RNA and SR6 are displayed. The Y axis refers to the relative amount of the corresponding RNA. The results of two or three independent experiments are shown.
The strongest effect was observed upon addition of ethanol. Within 0.5 min, yonT mRNA disappeared completely. By contrast, the amount of SR6 was only two-fold reduced after 1 min and increased to 80% of its initial amount after 5 min. In both cases, the effects were sigB independent.
Addition of vancomycin (cell-wall stress) resulted in a five-fold decrease in the amount of SR6 after 20 min whereas the amount of yonT RNA decreased four-fold after 60 min. Both effects were sigB independent.
After heat shock (from 37 °C to 48 °C), a four-fold increase in the amount of yonT mRNA was observed whereas the amount of SR6 decreased four-fold after 60 min, both sigB independently. This is in stark contrast to the heat-shock effects on bsrG and bsrE toxin mRNAs, whose amounts both decreased 10-fold while the amounts of the cognate antitoxins SR4 and SR5 were unaffected [15,21].
Upon manganese stress, the amount of SR6 decreased four-fold after 60 min, whereas the amount of yonT RNA increased 3.5-fold after 10 min followed by a decrease after 20 min to the duplicate of the initial amount, both independent of sigB.
Lower effects were found upon iron limitation (two-fold decreased amounts of both SR6 and yonT RNA), oxidative stress (two-fold decrease of SR6, no significant effect on yonT RNA), and salt stress (two-fold decrease of yonT RNA, at the most two-fold effect on SR6). Interestingly, oxidative stress and iron limitation were the only stress conditions that depended on sigB.
Taken together, we can conclude that yonT/SR6 is a multistress-responsive type I TA system.
2.9. Homologues of the yonT, yoyJ and sr6 Are Present in Some Other Bacillus Species
An inspection of all currently published genomes for homologues to yonT, yoyJ and sr6 revealed DNA sequences of high similarity in Bacillus subtilis, Bacillus amyloliquefaciens XH7, Bacillus atrophaeus NRS 1221A, Bacillus methylotrophicus JJ-D34 and Bacillus velezensis CC09 (Figure 8). The yonT DNA sequence and the YonT aa sequence were highly conserved in all five strains, the latter displaying only a few aa exchanges. In addition, a protein BLAST revealed a highly similar YonT protein comprising 66 aa in B. nakamurai and a shortened, 37 aa homologue lacking the YonT C terminus in B. vallismortis. The sr6 gene was also highly conserved in the five species mentioned first. In all cases, an extended −10 box of psr6 was detectable and only the first transcribed nt showed a few variations. By contrast, only B. subtilis does encode the complete yoyJ ORF. In B. atrophaeus, B. methylotrophicus and B. velezensis, the ATG start codon and the following 11 codons of yoyJ are missing and the shortened ORF starting with the second ATG is not preceded by a RBS, while the B. amyloliquefaciens yoyJ ORF carries a premature stop codon after codon 12 making it in all these cases unlikely that a functional YoyJ protein is synthesized.
Figure 8.
Alignments of the sr6 DNA sequences and the YonT and YoyJ ORFs from different Bacillus species. BLASTn and BLASTp were applied. Above: Amino acid sequence alignments. Above B. subtilis sequence, below sequences from B. amyloliquefaciens, B. athrophaeus, B. methylotrophicus and B. velezensis. Below: DNA sequence alignment.
3. Discussion
3.1. yonT/SR6 Is a Type I TA System That Encodes Two Toxins Regulated by Different Mechanisms
In this study, we report on the second type I TA system from the B. subtilis chromosome that is encoded on the SPβ prophage. In contrast to the other type I TA systems investigated so far in B. subtilis, but also in other bacteria, it encodes one RNA antitoxin (SR6) that regulates two toxin genes, yonT and yoyJ, employing two different mechanisms. The 3′ end of the sr6 gene overlaps the 3′ end of the yonT gene by about 65 bp. Both RNAs interact by complementary base-pairing leading to the decay of yonT mRNA indicated by its 4-fold shorter half-life in the presence of SR6 (Figure 6A). RNase III is involved in this process: Durand et al. have shown that RNase III is in B. subtilis—in contrast to other bacterial species—essential because it is needed to protect this bacterium from the expression of two toxin genes, txpA and yonT encoded on two prophage-born type I TA systems, txpA/RatA on the skin prophage and yonT/sr6 on the SPβ prophage [11]. Only in the presence of RNase III, the cognate antitoxins RatA and antisense-yonT (which we renamed as SR6) can prevent toxin synthesis [11]. Once both gene pairs are deleted, RNase III is not essential anymore. The mechanism of action of SR6 on yonT mRNA is similar to that of SR4 on bsrG and SR5 on bsrE mRNA [15,21]. In both of these cases, antitoxin binding to toxin mRNA recruits RNase III to cleave the duplex, in bsrG RNA at nt 14 from the 3′ end, in bsrE RNA 7-9 nt from the 3′ end. In contrast to SR5, SR4 is a bifunctional antitoxin as it does not only promote bsrG mRNA degradation but also impedes its translation by inducing a structural change that obscures the bsrG RBS [23]. As with translational yonT-lacZ fusions, no differences in translational activities could be observed in the absence or presence of SR6 (Table S1), a translation inhibition of yonT by SR6 is improbable. However, the measured β-galactosidase activities were only slightly above the negative control. Therefore, other experiments are required to corroborate these results.
Whereas SR6 acts like SR4 and SR5 by binding to the 3′ end of the toxin yonT RNA to promote its degradation, it overlaps the 5′ region of the second toxin gene, yoyJ, by approximately 30–40 bp. As we demonstrate in Figure 6, SR6 does not induce degradation of yoyJ mRNA. Unfortunately, employing a translational yoyJ-lacZ fusion under control of the strong promoter pIII [25] we could only measure activities barely above the levels of the empty vector control (Table S1), both in logarithmic or early stationary phase in the presence (strain DB104) and absence (strain 1A100) of SR6. Therefore, we resorted to in vitro yoyJ translation to analyze a possible effect of SR6. However, no in vitro translation product of yoyJ could be detected. The reason is the short distance (4 nt) between SD sequence and yoyJ start codon that makes yoyJ translation extremely inefficient (see Figure 2C).
However, transformation experiments provided an indirect evidence for the toxicity of YoyJ: B. subtilis strain A100 devoid of the yonT/yoyJ/SR6 system could not be transformed by plasmid pUCBYJ for constitutive yoyJ expression, whereas DB104 encoding SR6 as single copy on its chromosome, could, albeit with a somewhat lower efficiency than the same vector encoding yonU or sr6 itself (Figure 5). Although this data is only an indirect evidence, it shows that yoyJ does encode a toxin, albeit one with considerable lower toxicity than YonT. The low toxicity of YoyJ might be due to its very weak translation. For YonT it was not possible to construct an inducible overexpression E. coli-B. subtilis shuttle vector even with a medium copy number and a tightly controlled promoter. This was in contrast to a former publication that reported the construction of an E. coli plasmid for inducible overexpression of yonT under the pBAD promoter [6]. We assume that a mutation occurred either in the constructed plasmid or the E. coli strain that decreased toxicity of YonT and allowed to establish this plasmid in E. coli.
The growth and establishment effects suggest a slightly toxic function of the yoyJ gene. The antitoxin SR6 carries a complementary region to the 5′ end of yoyJ which is uncommon in type I TA systems of Gram-positive bacteria, but a characteristic of Gram-negative bacterial type I TA systems such as hok/Sok or ibs/Sib of E. coli (rev. in [4]). At the moment, the molecular mechanism by which SR6 neutralizes the toxicity of YoyJ is unknown. A BLAST search showed that yonT is well conserved in five Bacillus species, among them B. amyloliquefaciens and B. atrophaeus (Figure 8). By contrast, the yoyJ gene carries different mutations in other Bacillus species, e.g., an early stop codon in B. amyloliquefaciens and a mutated start codon in the other three species which indicates that YoyJ is not synthesized in other species.
3.2. Antitoxin/toxin RNA Ratios and Half-Lives
In cis-encoded sense/antisense RNA systems—to which type I TA systems belong—the amount of the antisense RNA has to be 5- to 10-fold higher than that of the sense RNA to ensure proper control (rev. in [26]). This has been also confirmed by quantitative measurements for type I TA systems bsrG/SR4 [27] and bsrE/SR5 [21]. In complex TY medium, the SR4/bsrG RNA ratio fell below 1.0 already at late log phase and decreased further drastically in stationary phase to 0.2 [27]. Similar results were obtained for bsrE/SR5 in the stationary growth phase in TY medium and in minimal CSE medium [21]. Furthermore, only under two stress conditions, anaerobic and alkaline stress, the SR5/bsrE RNA ratio fell well below 1, thus allowing toxin expression [21]. Here, we determined SR6/yonT mRNA ratios between 82 and 574 in TY medium and between 9 and 201 in CSE medium without glucose (Figure 3) excluding toxin expression under these conditions. By contrast, in CSE medium with glucose, this value fell already at late logarithmic phase below 2, indicating that yonT might be expressed under these conditions. However, it has to be taken into consideration that yonT has the rare GUG start codon preceded by an extended SD sequence with 11 bp complementarity to the anti-SD of B. subtilis 16S rRNA, which would efficiently recruit, but slowly release ribosomes [28], thus drastically reducing the probability of toxin synthesis [20].
A hallmark of all TA systems is a stable toxin regulated by an unstable antitoxin. This was confirmed by half-life measurements in several type I TA systems that revealed short half-lives for the regulatory RNA antitoxin and long half-lives for toxin mRNAs (rev. in [4]). In hok/Sok, the hok mRNA is extremely stable with a half-life of about 30 min, whereas antitoxin Sok is short-lived with a half-life of 1 min. Similarly, bsrG mRNA and bsrE mRNA have a half-life of 16 min and 80–120 min, respectively, whereas those of their cognate antitoxins SR4 and SR5 are 3.5 min and 7–14 min [15,21]. Surprisingly, the half-live of SR6 (both species) is with >60 min extremely long, whereas that of yonT mRNA is with 8 min more than 7 times shorter. As the psr6 activity is extremely low (see Figure 4), such a long half-life of SR6 might be required to ensure sufficient antitoxin levels. Due to its long half-life, the identification of RNases responsible for SR6 degradation is not feasible. Interestingly, Durand et al. postulated a half-life of ≈1.3 min for the yonT RNA in the presence of SR6 and a half-life of SR6 of >20 min [11]. However, they used another B. subtilis strain which might differ in the activity of certain RNases. In addition, their yonT RNA signal was barely visible making this half-life determination questionable. A determination of half-lives in the absence of SR6 was not performed by these authors. The second toxin RNA regulated by SR6, yoyJ-mRNA, is with a half-life of >90 min also very stable (Figure 6). Such a long half-life is rather typical for a toxin mRNA. In addition, in TY medium, but not in CSE or CSEG, two SR6 species exist, one with the expected length of about 100 nt, and a longer species of ≈215 nt (see Figure 1) that resulted from read-through of the apparently inefficient transcription terminator. The latter species was only detectable at transition phase. Currently, it is unclear which factors allow read-through from psr6 only in TY medium and during a short time-frame.
3.3. yonT/SR6 Is a Multistress-Responsive TA System
Toxin expression has to be tightly controlled: For PSK systems like hok/Sok and fst/RNAII, constitutive expression of both toxin and antitoxin promoters are required (rev. in [4]). The same seems to be true for chromosome-encoded txpA/RatA [10]. By contrast, B. subtilis bsrG and bsrE are temperature-sensitive [15,21]: bsrG mRNA has a 3- to 4-fold shorter half-life at high temperatures due to refolding at heat-shock which allows RNases Y and J1 to access newly formed single-stranded regions [27]. Furthermore, for the bsrE/SR5 system, multiple stress conditions affected toxin and/or antitoxin expression, the most pronounced among them were anaerobic and alkaline stress that decreased the antitoxin/toxin RNA ratio 8- to 10-fold and 4- to 5-fold, respectively, as well as ethanol stress, which caused a rapid degradation of toxin RNA by RNase Y within 0.5 min [21]. E. coli toxin tisB, symE and dinQ promoters are preceded by LexA boxes which make them SOS inducible whereas their antitoxins are expressed constitutively (rev. in [13,16]). In the case of SymE, an additional post-translational regulation by Lon protease was reported [16]. S. aureus sprA1 expression decreases two-fold at lower pH, while it increases three-fold under oxidative stress [29]. Interestingly, E. coli ralR is higher expressed in later stages of biofilm development [30]. Likewise, chromosome-encoded type II TA systems as relBE or mazEF were discovered to be sensitive to nutritional stress during growth (rev. in [4]).
Here, we report the effects of several stress factors on yonT mRNA and SR6 (see Figure 7). As in the case of bsrE mRNA, the strongest effect was found for ethanol: within 0.5 min after ethanol addition, yonT mRNA disappeared completely. In contrast to bsrE/SR5, oxygen deficiency did not have significant effects. However, the cell-wall toxin vancomycin, that did neither impact bsrE RNA nor SR5, resulted in a four- and five-fold decrease of yonT RNA and SR6, respectively. This would, however, not alter the SR6/yonT ratio making toxin expression under cell-wall stress unlikely. Interestingly, oxidative stress and iron limitation which resulted in about 2-fold effects on yonT RNA and SR6, were the only stress conditions dependent on sigB. In the case of bsrE/SR5, iron limitation increased the amount of antitoxin SR5, also sigB dependent, but oxidative stress had no effect. However, neither in bsrE/SR5 nor yonT/SR6 an additional sigB promoter could be detected, so that SigB dependency is most likely due to other factors controlled by this alternative sigma factor. Manganese stress had no effect on bsrE/SR5, but affected yonT/SR6 significantly, and salt stress had a small impact on yonT/SR6 while it exerted no effect on bsrE/SR5. Surprisingly, temperature shock had the reverse effect on yonT/SR6 compared to bsrE/SR5 or bsrG/SR4 [15,21]: It caused a four-fold increase in the amount of yonT mRNA whereas that of SR6 was reduced four-fold after 60 min. By contrast, bsrG and bsrE mRNA amounts decreased 10-fold after heat-shock while the amounts of both antitoxins remained constant. Based on these observations, it can be assumed that the toxin YonT might be synthesized upon heat shock. Since secondary structures of yonT RNA or SR6 have not been mapped so far, we currently cannot provide a mechanistic explanation for the heat-shock effects on the amounts of SR6 and yonT RNA.
In summary, we can conclude that yonT/SR6 is also a multistress-responsive type I TA system, and that most probably yonT is expressed upon heat-shock conditions at least in individual cells of the culture.
Future investigations will focus on the elucidation of the mechanisms responsible for the stress effects and on the cellular targets of the two toxins, YonT and YoyJ.
4. Materials and Methods
4.1. Enzymes and DNA Manipulations
Taq DNA polymerase (Roche), Firepol polymerase (Solis Biodyne), sequenase and Thermoscript reverse transcriptase (Thermoscientific), as well as T7 polymerase and polynucleotide kinase (New England Biolabs) were used. DNA manipulations like plasmid purification, E. coli and B. subtilis transformation were carried out as described [31].
4.2. Strains, Media and Isolation of Chromosomal DNA
E. coli strain DH5α and B. subtilis strains DB104 and 1A100 (ΔSPβ) were used (see Table 1). 1A100 was included as this strain does not carry the yonT/sr6 region encoded on the SPβ prophage. TY served as complex medium for E. coli and B. subtilis and CSE as minimal medium for B. subtilis [32]. Chromosomal DNA from B. subtilis strains was isolated as described [33].
Table 1.
Bacterial strains.
4.3. Primer Extension
Primer extension experiments were carried out as described [33] using total RNA from B. subtilis strain DB104 and 5′-labelled primer SB2646 for yonT RNA, SB2648 for SR6 and SB2647 for yonU RNA (primers are listed in Table S2).
4.4. Vector Construction
For the determination of promoter activities, transcriptional lacZ fusion plasmids were constructed as follows: A PCR was performed on chromosomal DNA of B. subtilis DB104 with primer pairs SB2598/2599 (pMGCR1/5), SB2628/2599 (plasmid pMGCR6), SB2630/2644 (pMGCR8), SB2600/SB264 (pMGCR14) and SB2630/SB2706 (pMGCR16). All primers are listed in Table S2, and all plasmids in Table 2. The resulting fragments were cleaved with BamHI and EcoRI—except pMGCR1/5 that was cleaved with BglII and BamHI—and inserted into the pMG16 vector digested with the same enzyme pair. Plasmids pMGCR1/5 and pMGCR6 contain pyonT from −500 and −100, respectively, to +10 with regard to the transcriptional start site (TSS) fused to the promoterless lacZ gene, whereas pMGCR14 comprises psr6 with −500 to +50 and pMGCR8 and pMGCR16 psr6 with −240 to +50, respectively. Translational yonT-lacZ (pGAY1) and yoyJ-lacZ (pGAY2) fusions were constructed using a PCR with primer pairs SB2760/2761 (pGAY1) or SB2764/SB2765 (pGAY2), respectively on chromosomal DNA. The resulting fragments were digested with BamHI and EcoRI and inserted into the BamH/EcoRI pGAB1 vector. In all cases, the native promoter was replaced by the strong constitutive heterologous promoter pIII [25].
Table 2.
Plasmids used in this study.
| Plasmid | Description | Reference |
|---|---|---|
| pUCB2 | Shuttle vector of pUC19 and pUB110, NeoR, PhleoR | [38] |
| pDR111 | Vector for IPTG-inducible overexpression and integration into the B. subtilis amyE locus, SpecR | [37] |
| pAPNC213cat | Vector for IPTG-inducible overexpression and integration into the B. subtilis AP locus, CmR | [24] |
| pMG16 | Vector for integration of transcriptional lacZ fusions into B. subtilis amyE locus, SpecR | M. Gimpel, unpublished |
| pGAB1 | Vector for integration of translational lacZ fusions into B. subtilis amyE locus, KanR | S. Brantl, unpublished |
| pMGCR1 | pMG16 with yonT from −500 to +10 * | This study |
| pMGCR6 | pMG16 with yonT from −100 to +10 * | This study |
| pMGCR8 | pMG16 with sr6 from −240 to +50 * | This study |
| pMGCR14 | pMG16 with sr6 from −500 to +50 * | This study |
| pGAY1 | pGAB1 with yonT gene | This study |
| pGAY2 | pGAB1 with yoyJ gene | This study |
| pUCBas | pUCB2 with sr6 gene | This study |
| pUCBYT | pUCB2 with yonT gene | This study |
| pUCBYJ | pUCB2 with yoyJ gene | This study |
| pUCBYU | pUCB2 with yonU gene | This study |
| pAPYT3 | pAPNC213cat with yonT gene (M1Stop) | This study |
| pDRYJ | pDR111 with yoyJ gene | This study |
* with regard to the TSS (transcription start site).
To overexpress sr6, yoyJ and yonU in Bacillus subtilis, plasmids pUCBas, pUCBYJ and pUCBYU were constructed, respectively, as follows: PCR fragments were obtained on chromosomal DNA from B. subtilis strain DB104 with primer pairs SB2654/2655 (pUCBas), SB2690/2691 (pUCBYJ) or SB2635/SB2636 (pUCBYU), cleaved with BamHI and HindIII and inserted into the BamHI/HindIII vector of pUCB2. The cognate psr6 and pyonU promoters and, in case of pUCBYJ, pyonT, were used. Primer SB2636 contains the heterologous bsrF terminator at its 3′ end [36]. pANC213cat is a vector for integration into the alkaline phosphatase gene (aprE gene) that comprises the tightly controlled IPTG-inducible pSPAC promoter. A promoterless yonT fragment generated on chromosomal DNA as template with primer pair SB2660/SB2661 was cleaved with BamHI and EcoRI and inserted into the BamHI/EcoRI pANC213cat vector. In the resulting plasmid pAPYT3, the yonT start codon was replaced by a stop codon and the bsrF terminator added at the 3′ end. A promoterless yoyJ fragment was produced by PCR on genomic DNA with primer pair SB2722/SB2723, digested with SphI and HindIII and cloned into the SphI/HindIII pDR111 [37] vector that contains the IPTG-inducible hyperspank promoter and can be integrated into the B. subtilis amyE locus.
4.5. β-Galactosidase Assay
The transcriptional and translational fusions were integrated into the amyE locus of the B. subtilis DB104 or 1A100 chromosome and β-galactosidase activities were determined as described [38].
4.6. Determination of the Intracellular Concentrations of yonT mRNA, yoyJ mRNA and SR6
The intracellular amounts of the RNAs were determined as described previously [21].
4.7. Preparation of total RNA, RNA Gel Electrophoresis and Northern Blotting
Preparation of total RNA, RNA gel electrophoresis on 6% denaturing polyacrylamide gels, and Northern blotting were carried out as described previously [33]. For the detection of yonT mRNA, yoyJ mRNA, yonU mRNA and SR6, [α-32P] UTP-labeled riboprobes were used generated with T7 RNA polymerase on PCR-derived DNA fragments as described [39]. RNA half-life determinations were performed according to [40], except that 200 μg/mL rifampicin were used.
4.8. Stress Conditions
The following stress conditions were applied (final concentrations are indicated): ethanol stress (4% ethanol), oxidative stress (10 mM H2O2), salt stress (0.5 M NaCl), Mn2+ stress (1 mM MnCl2), iron depletion (250 μM 2,2-Dipyridyl-N,N-dimethylsemicarbazone = DIP, dissolved in DMSO) and cell wall antibiotic stress (10 and 20 μg/mL vancomycin). For heat shock, cultures were shocked from 37 °C to 48 °C.
Acknowledgments
We are very grateful to Natalie Jahn (formerly AG Bakteriengenetik, Jena) for her help with the adaptation of the experimental methods to the yonT/SR6 system. Furthermore, we thank Peter Müller (AG Bakteriengenetik, Jena) for the search for yonT/sr6/yoyJ genes in other bacterial species and for critical reading of the manuscript. This work was supported by Deutsche Forschungsgemeinschaft BR1552/10-1 (to S.B.).
Abbreviations
The following abbreviations are used in this manuscript:
aa, amino acid; nt, nucleotide; bp, base pair; sRNA, small RNA, RBS, ribosome binding site, TA, toxin-antitoxin.
Supplementary Materials
The following are available online at http://www.mdpi.com/2072-6651/10/2/74/s1, Figure S1: RT PCR on gapA RNA, Figure S2: Calculation of the amounts of yonT RNA and SR6 using standard curves (shown above) obtained by loading and Northern blotting of defined amounts of in vitro synthesized yonT RNAand SR6 in the range of 0.01 to 5 fmol in parallel onto the gels with RNA samples from cultures grown in TY, CSEG and CSE medium (see Figure 3 in the main body of publication), Table S1: β-galactosidase-activities of translational yonT-1 lacZ and yoyJ lacZ fusions, Table S2: Oligonucleotides used in this study.
Author Contributions
S.B. conceived and designed the experiments, C.R. and C.L. performed the experiments; C.R., C.L. and S.B. analyzed the data; S.B. wrote the paper.
Conflicts of Interest
No potential conflicts of interest were disclosed.
Key Contribution
The yonT/yoyJ/SR6 system is the first type I TA system in which the antitoxin regulates two toxins using two different mechanisms, promotion of RNA degradation and inhibition of translation.
References
- 1.Brantl S. Bacterial type I toxin-antitoxin systems. RNA Biol. 2012;9:1488–1490. doi: 10.4161/rna.23045. [DOI] [PubMed] [Google Scholar]
- 2.Page R., Peti W. Toxin-antitoxin systems in bacterial growth arrest and persistence. Nat. Chem. Biol. 2016;12:208–214. doi: 10.1038/nchembio.2044. [DOI] [PubMed] [Google Scholar]
- 3.Aakre C.D., Phung T.N., Huang D., Laub M.T. A bacterial toxin inhibits DNA replication elongation through a direct interaction with the β sliding clamp. Mol. Cell. 2013;52:617–628. doi: 10.1016/j.molcel.2013.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brantl S., Jahn N. sRNAs in bacterial type I and type III toxin/antitoxin systems. FEMS Microbiol. Rev. 2015;39:413–427. doi: 10.1093/femsre/fuv003. [DOI] [PubMed] [Google Scholar]
- 5.Gerdes K., Wagner E.G.H. RNA antitoxins. Curr. Opin. Microbiol. 2007;10:117–124. doi: 10.1016/j.mib.2007.03.003. [DOI] [PubMed] [Google Scholar]
- 6.Fozo E.M., Makarova K.S., Shabalina S.A., Yutin N., Koonin E.V., Storz G. Abundance of type I toxin-antitoxin systems in bacteria: Searches for new candidates and discovery of novel families. Nucleic Acids Res. 2010;38:3743–3759. doi: 10.1093/nar/gkq054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Weaver K.E. The par toxin-antitoxin system from Enterococcus faecalis plasmid pAD1 and its chromosomal homologs. RNA Biol. 2012;9:1498–1503. doi: 10.4161/rna.22311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Weaver K.E. The type I toxin-antitoxin par locus from Enterococcus faecalis plasmid pAD1: RNA regulation by both cis- and trans-acting elements. Plasmid. 2014;78:65–70. doi: 10.1016/j.plasmid.2014.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gerdes K., Christensen S.K., Løbner-Olesen A. Prokaryotic toxin-antitoxin stress response loci. Nat. Rev. Microbiol. 2005;3:371–382. doi: 10.1038/nrmicro1147. [DOI] [PubMed] [Google Scholar]
- 10.Silvaggi J.M., Perkins J.B., Losick R. Small untranslated RNA antitoxin in Bacillus subtilis. J. Bacteriol. 2005;187:6641–6650. doi: 10.1128/JB.187.19.6641-6650.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Durand S., Gilet L., Condon C. The essential function of B. subtilis RNase III is to silence foreign toxic genes. PLoS Genet. 2012;8:e1003181. doi: 10.1371/journal.pgen.1003181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Arnion H., Korkut D.N., Masachis Gelo S., Chabas S., Reignier J., Iost I., Darfeuille F. Mechanistic insights into type I toxin antitoxin systems in Helicobacter pylori: The importance of mRNA folding in controlling toxin expression. Nucleic Acids Res. 2017;45:4782–4795. doi: 10.1093/nar/gkw1343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wagner E.G.H., Unoson C. The toxin-antitoxin system tisB-istR1. Expression, regulation and biological role in persister phenotypes. RNA Biol. 2012;9:1513–1519. doi: 10.4161/rna.22578. [DOI] [PubMed] [Google Scholar]
- 14.Fozo E.M. New type I toxin-antitoxin families from “wild” and laboratory strains of E. coli. Ibs-Sib, ShoB-OhsC and Zor-Orz. RNA Biol. 2012;9:1504–1512. doi: 10.4161/rna.22568. [DOI] [PubMed] [Google Scholar]
- 15.Jahn N., Preis H., Wiedemann C., Brantl S. BsrG/SR4 from Bacillus subtilis—The first temperature-dependent type I toxin-antitoxin system. Mol. Microbiol. 2012;83:579–598. doi: 10.1111/j.1365-2958.2011.07952.x. [DOI] [PubMed] [Google Scholar]
- 16.Kawano M. Divergently overlapping cis-encoded antisense RNA regulating toxin-antitoxin systems from E. coli: Hok/sok, ldr/rdl, symE/symR. RNA Biol. 2012;9:1520–1527. doi: 10.4161/rna.22757. [DOI] [PubMed] [Google Scholar]
- 17.Weel-Sneve R., Kristiansen K., Odsbu I., Dalhus B., Booth J., Rognes T., Skarstad K., Bjoras M. Single transmembrane peptide DinQ modulates membrane-dependent activities. PLoS Genet. 2013;8:e1003260. doi: 10.1371/journal.pgen.1003260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Guo Y., Quiroga C., Chen Q., McAnulty M.J., Benedik M.J., Wood T.K., Wang X. RalR (a DNase) and RalA (a small RNA) form a type I toxin-antitoxin system in Escherichia coli. Nucleic Acids Res. 2014;42:6448–6462. doi: 10.1093/nar/gku279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Berghoff B.A., Hoekzema M., Aulbach L., Wagner E.G.H. Two regulatory RNA elements affect TisB-dependent depolarization and persister formation. Mol. Microbiol. 2017;103:1020–1033. doi: 10.1111/mmi.13607. [DOI] [PubMed] [Google Scholar]
- 20.Durand S., Jahn N., Condon C., Brantl S. Type I toxin-antitoxin systems in Bacillus subtilis. RNA Biol. 2012;9:1491–1497. doi: 10.4161/rna.22358. [DOI] [PubMed] [Google Scholar]
- 21.Müller P., Jahn N., Ring C., Maiwald C., Neubert R., Meißner C., Brantl S. A multistress responsive type I toxin-antitoxin system: BsrE/SR5 from the B. subtilis chromosome. RNA Biol. 2016;13:511–523. doi: 10.1080/15476286.2016.1156288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Meißner C., Jahn N., Brantl S. In vitro characterization of the type I toxin-antitoxin system bsrE/SR5 from Bacillus subtilis. J. Biol. Chem. 2016;291:560–571. doi: 10.1074/jbc.M115.697524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jahn N., Brantl S. One antitoxin—two functions: SR4 controls toxin mRNA decay and translation. Nucleic Acids Res. 2013;41:9870–9880. doi: 10.1093/nar/gkt735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jahn N., Brantl S., Strahl H. Against the mainstream: The membrane associated type I toxin BsrG from Bacillus subtilis interferes with cell envelope biosynthesis without increasing membrane permeability. Mol. Microbiol. 2015;98:651–666. doi: 10.1111/mmi.13146. [DOI] [PubMed] [Google Scholar]
- 25.Brantl S. The copR gene product of plasmid pIP501 acts as a transcriptional repressor at the essential repR promoter. Mol. Microbiol. 1994;14:473–483. doi: 10.1111/j.1365-2958.1994.tb02182.x. [DOI] [PubMed] [Google Scholar]
- 26.Brantl S. Regulatory mechanisms employed by cis-encoded antisense RNAs. Curr. Opin. Microbiol. 2007;10:102–109. doi: 10.1016/j.mib.2007.03.012. [DOI] [PubMed] [Google Scholar]
- 27.Jahn N., Brantl S. Heat shock induced refolding entails rapid degradation of bsrG toxin mRNA by RNases Y and J1. Microbiology. 2016;162:590–599. doi: 10.1099/mic.0.000247. [DOI] [PubMed] [Google Scholar]
- 28.Li G.W., Oh E., Weissman J.S. The anti-Shine-Dalgarno sequence drives translational pausing and codon choice in bacteria. Nature. 2012;484:538–541. doi: 10.1038/nature10965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sayed N., Jousselin A., Felden B. A cis-antisense RNA acts in trans in Staphylococcus aureus to control translation of a human cytolytic peptide. Nat. Struct. Mol. Biol. 2011;19:105–112. doi: 10.1038/nsmb.2193. [DOI] [PubMed] [Google Scholar]
- 30.Domka J., Lee J., Bansal T., Wood T.K. Temporal gene-expression in Escherichia coli K-12 biofilms. Environ. Microbiol. 2007;9:332–3346. doi: 10.1111/j.1462-2920.2006.01143.x. [DOI] [PubMed] [Google Scholar]
- 31.Brantl S., Behnke D. Characterization of the minimal origin required for replication of the streptococcal plasmid pIP501 in Bacillus subtilis. Mol. Microbiol. 1992;6:3501–3510. doi: 10.1111/j.1365-2958.1992.tb01785.x. [DOI] [PubMed] [Google Scholar]
- 32.Heidrich N., Chinali A., Gerth U., Brantl S. The small untranslated RNA SR1 from the B. subtilis genome is involved in the regulation of arginine catabolism. Mol. Microbiol. 2006;62:520–536. doi: 10.1111/j.1365-2958.2006.05384.x. [DOI] [PubMed] [Google Scholar]
- 33.Licht A., Preis S., Brantl S. Implication of CcpN in the regulation of a novel untranslated RNA (SR1) in B. subtilis. Mol. Microbiol. 2005;58:189–206. doi: 10.1111/j.1365-2958.2005.04810.x. [DOI] [PubMed] [Google Scholar]
- 34.Hanahan D. Studies on transformation of Escherichia coli with plasmids. J. Mol. Biol. 1983;166:557–580. doi: 10.1016/S0022-2836(83)80284-8. [DOI] [PubMed] [Google Scholar]
- 35.Kawamura F., Doi R.H. Construction of a Bacillus subtilis double mutant deficient in extracellular alkaline and neutral proteases. J. Bacteriol. 1984;160:442–444. doi: 10.1128/jb.160.1.442-444.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Preis H., Eckart R.A., Gudipati R.K., Brantl S. CodY activates transcription of a small RNA in Bacillus subtilis. J. Bacteriol. 2009;191:5446–5457. doi: 10.1128/JB.00602-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Van Ooij C., Losick R. Subcellular localization of a small sporulation protein in Bacillus subtilis. J. Bacteriol. 2003;185:1391–1398. doi: 10.1128/JB.185.4.1391-1398.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gimpel M., Heidrich N., Mäder U., Krügel H., Brantl S. A dual-function sRNA from Bacillus subtilis: SR1 acts as a peptide-encoding mRNA on the gapA operon. Mol. Microbiol. 2010;76:990–1009. doi: 10.1111/j.1365-2958.2010.07158.x. [DOI] [PubMed] [Google Scholar]
- 39.Heidrich N., Brantl S. Antisense-RNA mediated transcriptional attenuation: Importance of a U-turn loop structure in the target RNA of plasmid pIP501 for the efficient inhibition by the antisense RNA. J. Mol. Biol. 2003;333:917–929. doi: 10.1016/j.jmb.2003.09.020. [DOI] [PubMed] [Google Scholar]
- 40.Brantl S., Wagner E.G.H. An unusually long-lived antisense RNA in plasmid copy number control: In vivo RNAs encoded by the streptococcal plasmid pIP501. J. Mol. Biol. 1996;255:275–288. doi: 10.1006/jmbi.1996.0023. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.