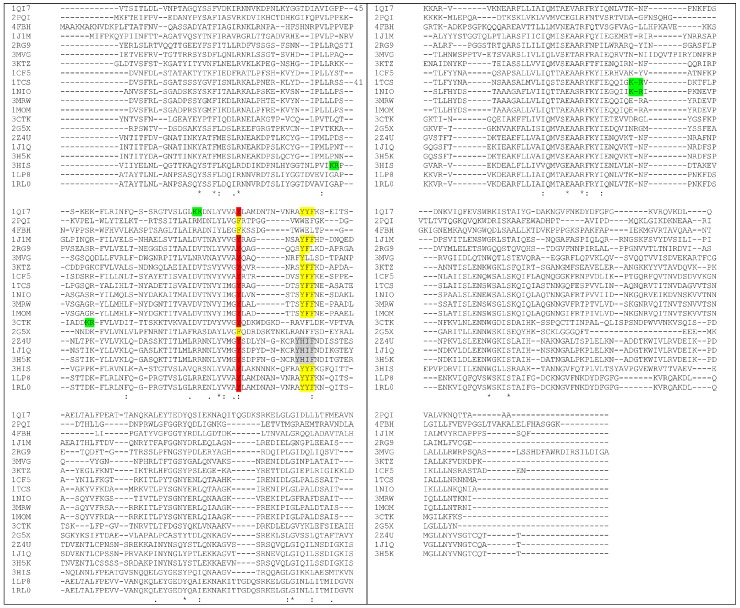
Figure 4.
Sequence alignment of RIPs with known 3D structure (each one is indicated by the corresponding PDB code). YFF or similar motifs are shown in yellow. KR motif is shown in green. Tyrosine 76 (numbers refer to Saporin sequence, 1QI7) that can have a role in maintaining Y88 in position (see also below) is shown in red. Only three RIPs have F in this position. Alignment has been performed using CLUSTAL O(1.2.4) software [65]. PDB codes correspond to: 1QI7 Saporin SO6 from Saponaria officinalis seeds 2PQI RIP from maize; 4FBH RIP from barley seeds; 1J1M Ricin A-Chain; 3MVG RIP Iris hollandica; 3KTZ GAP31; 1CF5 β-Momorcharin from Momordica charantia; 1TCS Tricosanthin from Trichosanthes kirilowii; 1NIO β-Luffin from Luffa cylindrica; 1MOM Momordin from Momordica charantia; 3CTK Bouganin Bougainvillea spectabilis; 2G5X Lychnin from Lychnis chalcedonica; 2Z4U PD-L4 from Phytolacca dioica; 1J1Q Pokeweed Antiviral Protein from Seeds (PAP-S1) from Phytolacca americana; 3H5K PD-L1 from Phytolacca dioica; 3HIS Saporin-L1 from Saponaria officinalis leaves; 1LP8 Dianthin from Dianthus caryophyllus.