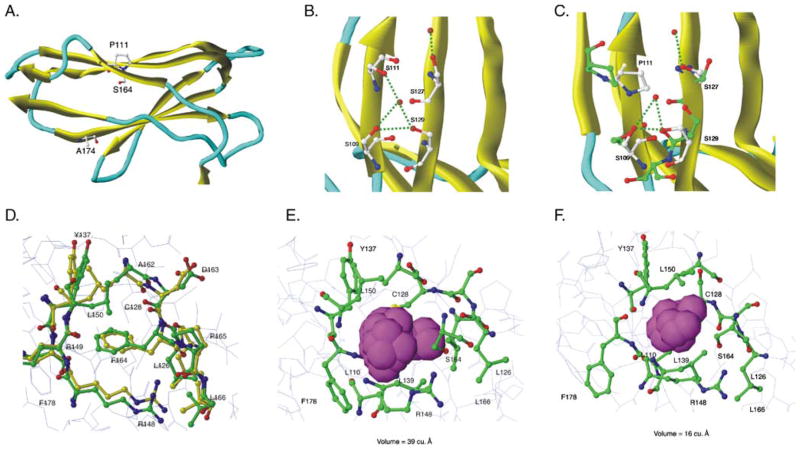
Fig. 5.
Molecular modeling predicts that P111 and S164 cause extracellular domain instability. KIR2DS5 (*001 and *002) energy-minimized structures were generated from the previously-solved KIR2DS2 crystal structure of extracellular domains (PDB:1M4K). a The D2 domain of KIR2DS5 (*001) showing the positions of the three polymorphic amino acids that differ between the receptors encoded by KIR2DS5*001 (shown) and KIR2DS5*002 (S111, F164, T174). Residues 111 and 174 lie on the surface; residue 164 lies in the hydrophobic interior of the domain. b Initial predicted KIR2DS5 (*002) structure in the region of S111 showing a hydrogen bond network with nearby serines. MD simulations did not alter this network (data not shown). c Predicted KIR2DS5 (*001) structure in the region of P111 before (yellow) and after (green) MD simulations. The substitution of proline for serine at 111 alters the hydrogen bond network destabilizing the structure. d Initial predicted KIR2DS5 (*002) structure before and after MD simulations. Yellow represents carbon atoms before MD simulations and green, after simulations. Red represents oxygen and blue, nitrogen. F164 interacts with hydrophobic residues in the interior of the domain and these interactions are not altered following MD simulations. e A closer view of the 2DS5*001-encoded S164 region before MD simulations. The van der Waals spheres are represented by a space filling model with magenta representing the pocket size in the S164 region. The size of the pocket before MD simulations was 39 Å3. f A closer view of 2DS5 (*001) S164 region after MD simulations. Magenta van der Waals spheres represent the volume of the pocket after MD simulations; its size was 16 Å3