Abstract
Here, we report that CRISPR guide RNAs (gRNAs) with a 5′-triphosphate group (5′-ppp gRNAs) produced via in vitro transcription trigger RNA-sensing innate immune responses in human and murine cells, leading to cytotoxicity. 5′-ppp gRNAs in the cytosol are recognized by DDX58, which in turn activates type I interferon responses, causing up to ∼80% cell death. We show that the triphosphate group can be removed by a phosphatase in vitro and that the resulting 5′-hydroxyl gRNAs in complex with Cas9 or Cpf1 avoid innate immune responses and can achieve targeted mutagenesis at a frequency of 95% in primary human CD4+ T cells. These results are in line with previous findings that chemically synthesized sgRNAs with a 5′-hydroxyl group are much more efficient than in vitro–transcribed (IVT) sgRNAs in human and other mammalian cells. The phosphatase treatment of IVT sgRNAs is a cost-effective method for making highly active sgRNAs, avoiding innate immune responses in human cells.
Clustered, regularly interspaced, short palindromic repeat (CRISPR)-CRISPR associated (Cas) nucleases, which function as an adaptive immune system in bacteria and archaea, have been repurposed for targeted genome editing in various cells and organisms (Cho et al. 2013a; Kim and Kim 2014). The Cas9 nuclease, the protein component in a type II CRISPR-Cas system, is complexed with two small RNAs termed a target-specific CRISPR RNA (crRNA) (∼40 nucleotides [nt] in length) and a target-independent trans-activating crRNA (tracrRNA) (∼86-nt) to form active RNA-guided endonucleases (Deltcheva et al. 2011; Jinek et al. 2012), which cleave DNA in a targeted manner, leading to site-specific genome modifications. The essential parts of the crRNA and tracrRNA are usually combined to generate a ∼100-nt single guide RNA (sgRNA). Cpf1 is another RNA-guided programmable nuclease, which is derived from a class 2 type V CRISPR-Cas system (Zetsche et al. 2015). Unlike Cas9, Cpf1 requires only a single 42-nt crRNA to cleave target DNA.
For successful CRISPR-mediated genome editing, efficient delivery or high level expression of Cas9 or Cpf1 and a guide RNA (gRNA) is essential. Plasmids encoding Cas9 and a gRNA or in vitro–transcribed Cas9 mRNA and a gRNA can be codelivered into cells. As an alternative to these methods, preassembled Cas9 or Cpf1 ribonucleoproteins (RNPs), composed of the purified Cas9 or Cpf1 protein and a gRNA, can be also delivered into cells via microinjection (Cho et al. 2013b; Sung et al. 2014), electroporation (Kim et al. 2014, 2016; Hur et al. 2016), cationic lipid-mediated transfection (Zuris et al. 2015), or protein transduction (Ramakrishna et al. 2014). RNPs cut target DNA immediately and are turned over rapidly in cells, which can alleviate off-target effects (Kim et al. 2014) and reduce mosaicism in whole organisms (Woo et al. 2015). Unlike plasmid transfection, RNP delivery is not associated with any risk of foreign DNA integration into the host genome (Kim et al. 2014) and cannot trigger cyclic GMP-AMP synthase (cGAS) activation, an innate immune system in mammalian cells that protects against foreign DNA (Sun et al. 2013). This advantage of RNP delivery over plasmid transfection has made Cas9, Cpf1, and Base Editor RNPs broadly useful for biomedical research and biotechnology (Woo et al. 2015; Dever et al. 2016; Kim et al. 2017a,c; Liang et al. 2017).
gRNAs in RNP complexes are typically transcribed in vitro using phage RNA polymerases such as T7 and SP6 polymerases. These in vitro–transcribed (IVT) gRNAs contain a triphosphate group at the 5′ terminus, which can induce a type I interferon-mediated immune response in various cell types (Kim et al. 2004; Pichlmair et al. 2006). The nascent 5′-triphosphate moiety in such an RNA molecule is recognized as part of a non-self RNA derived from viral infection. Immune sensing of non-self nucleic acids by innate immune systems leads to the induction of the type I interferons (IFNs), which in turn activate genes that encode antiviral effector proteins such as DDX58 (DExD/H-box helicase 58, also known as RIG-I), OAS2 (2′-5′-oligoadenylate synthetase), and PKR (dsRNA-dependent protein kinase) (Schlee and Hartmann 2016).
Here, we investigate whether the 5′-triphosphate moiety in IVT gRNAs indeed triggers the RNA-sensing innate immune response in human cells, leading to cell death, and show how to avoid this problem.
Results
Activation of type I interferon responses by 5′-triphosphate gRNA
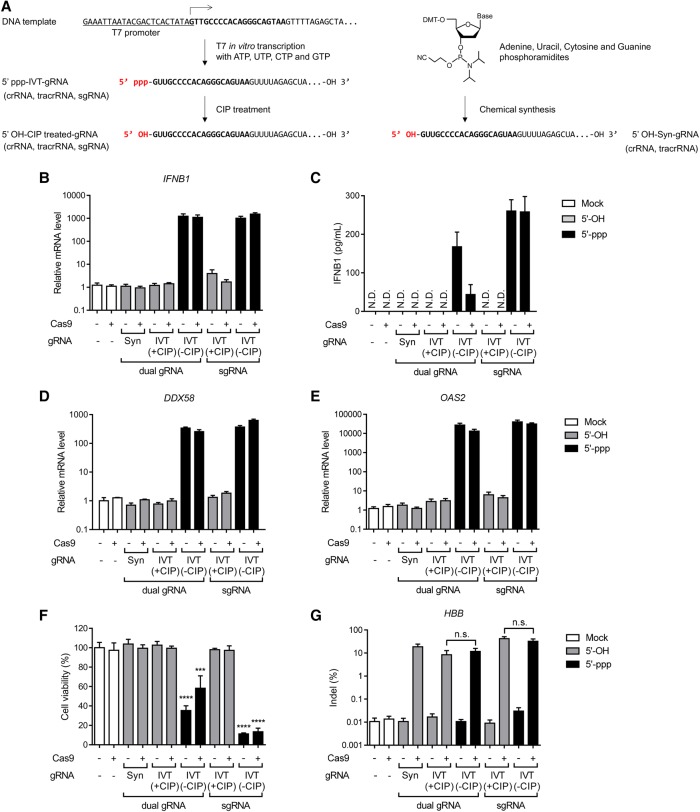
We first tested whether IVT gRNAs (dual RNA [crRNA plus tracrRNA] or sgRNA) could activate innate immune responses in cultured human cells. HBB gene-specific gRNAs were synthesized using T7 polymerase, complexed with the recombinant Streptococcus pyogenes Cas9 protein expressed in and purified from Escherichia coli and transfected into HeLa cells via lipofection (Fig. 1A). The expression of IFNB1, a hallmark of the type I IFN pathway, was evaluated by quantitative PCR and ELISA 24 h after transfection. The Cas9 RNP up-regulated IFNB1 mRNA levels by ∼1000-fold and protein levels by ∼200-fold (Fig. 1B,C). The Cas9 protein alone did not activate the IFN pathway, whereas 5′-triphosphorylated (5′-ppp) gRNAs alone activated the expression of IFNB1. In sharp contrast, transfection of 5′-hydroxylated (5′-OH) gRNAs, prepared by chemical synthesis or by treatment of IVT gRNAs with calf intestinal phosphatase (CIP), caused no significant difference in IFNB1 expression compared to that of untreated negative controls. IFN-stimulated DDX58 and OAS2 genes were also up-regulated by 5′-ppp gRNAs but not by 5′-OH gRNAs (Fig. 1D,E). Consistent with these results, cytotoxicity, caused by the type I IFN pathway, was observed with 5′-ppp sgRNAs but not with 5′-OH gRNAs (Fig. 1F). Thus, >80% of cells were killed by 5′-ppp sgRNAs. Dual RNAs containing the triphosphate group were also cytotoxic, killing up to 60% of transfected cells.
Figure 1.
Induction of type I interferon-mediated innate immune responses in HeLa cells by in vitro–transcribed gRNA containing 5′-triphosphate. (A) Schematic representation of gRNA 5′ ends, which vary according to the gRNA preparation method. gRNA sequences complementary to the HBB-target site are shown in bold. (B,D,E) Relative IFNB1 (B), DDX58 (D), and OAS2 (E) mRNA levels 24 h after transfection. Error bars, SEM; n = 3. (C) IFNB1 levels were measured by ELISA. Error bars, SEM; n = 3. (N.D.) None detected. (F) Cell viabilities after RNP transfection were determined by WST-1 assays. Error bars, SEM; n = 2. Statistical significances between the different measurements were calculated using one-way ANOVA. (***) P < 0.001, (****) P < 0.0001. (G) Indel frequencies induced by the HBB-targeting Cas9 RNP were measured by targeted deep sequencing. Error bars, SEM; n = 3. Statistical significances were calculated by t-test. (n.s.) Not significant, (Syn) chemically synthesized guide RNA, (IVT) in vitro–transcribed gRNA, (±CIP) in vitro–transcribed gRNA with or without CIP treatment.
Next, we measured HBB gene editing efficiencies in human cells transfected with 5′-ppp gRNAs or 5′-OH gRNAs (Fig. 1G). Both 5′-OH gRNAs and 5′-ppp gRNAs were equally efficient in HeLa cells, demonstrating that the triphosphate moiety at the 5′-end of sgRNAs does not affect DNA cleavage activity in human cells and suggesting that indels were formed before massive cell death. Note also that 5′-ppp gRNA transfection triggers production and secretion of interferon proteins in the media, which then bind to ubiquitously expressed receptors even in untransfected cells, leading to cell death.
We further investigated whether 5′-ppp-gRNAs trigger RNA-sensing immune responses in primary mouse embryonic fibroblast (MEF) cells (Supplemental Fig. S1A). The Ifnb1 and IFN-stimulated genes Ddx58 and Oas2 were up-regulated in MEF cells transfected with 5′-ppp-sgRNAs generated via in vitro transcription but not in those transfected with 5′-OH-gRNAs prepared by chemical synthesis or CIP treatment of IVT sgRNAs (Supplemental Fig. S1B–D). There was no difference in gene editing efficiencies between 5′-ppp-sgRNA and 5′-OH-sgRNA cotransfected with the Cas9 protein (Supplemental Fig. S1E).
RNA-sensing immune response to CRISPR-RNA via DDX58
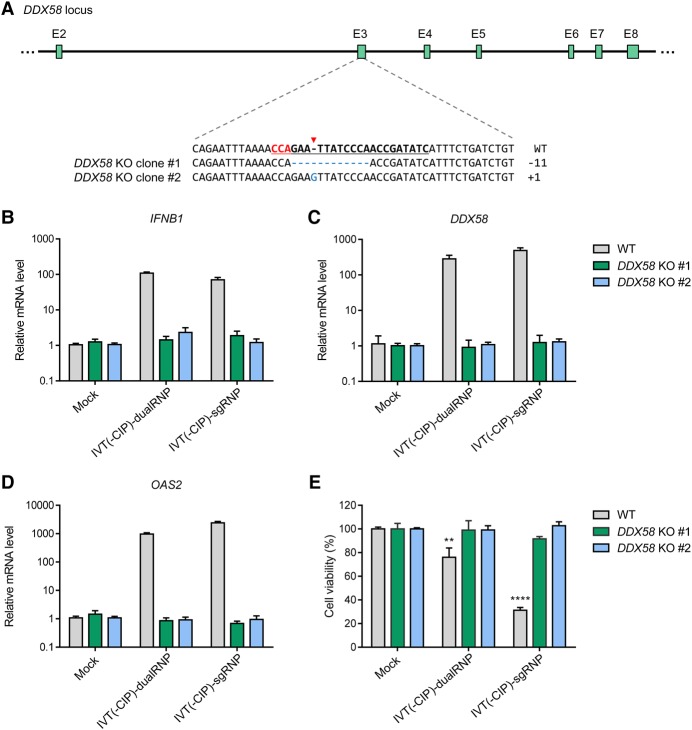
Cytosolic 5′-triphosphorylated RNAs, the main ligand for the DDX58 receptor, can function as an indicator of viral infection (Pichlmair et al. 2006; Rehwinkel et al. 2010). Hence, we hypothesized that DDX58 might be involved as the immune sensor responsible for detection of IVT gRNAs. To verify this, we first generated DDX58 knockout (KO) cell lines using a DDX58-specific sgRNA and Cas9 (Fig. 2A). We chose two KO clones with out-of-frame indel mutations. mRNA levels of IFNB1, DDX58, and OAS2 were not changed in these DDX58 KO cells transfected with 5′-ppp-gRNAs (Fig. 2B–D). Furthermore, IFN-mediated cytotoxicity associated with 5′-ppp-gRNAs was not detectably induced in these cells (Fig. 2E), suggesting that DDX58 is responsible for the innate immune response to IVT gRNAs with the 5′ triphosphate moiety.
Figure 2.
The type I interferon-mediated immune response triggered by gRNA is associated with DDX58. (A) Genotypes of DDX58 knockout (KO) HeLa cell lines. DDX58 KO clones #1 and #2 have homozygous out-of-frame mutations at the DDX58 locus. The target sequence complementary to the sgRNA used for mutagenesis is underlined and the PAM sequence is shown in red. (B–D) Relative IFNB1 (B), DDX58 (C), and OAS2 (D) mRNA levels in wild-type (WT) and DDX58 KO cell lines at 24 h after transfection. Error bars, SEM; n = 3. (E) WT and DDX58 KO cell viabilities after RNP transfection were determined by WST-1 assays. Error bars, SEM; n = 3. Statistical significances among the different measurements were calculated using one-way ANOVA. (**) P < 0.01, (****) P < 0.0001.
Type I interferon response induced by crRNA in Cpf1 RNPs
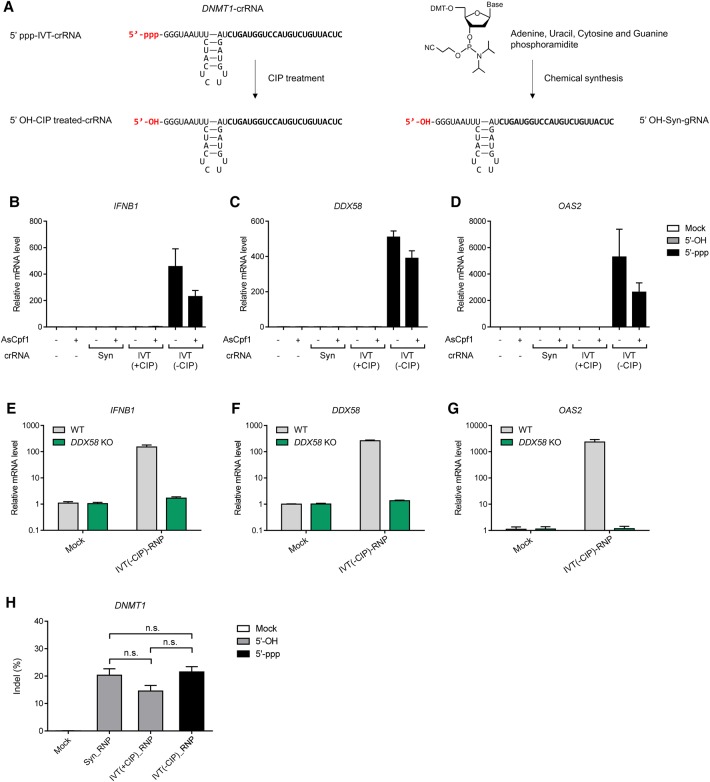
Next, we investigated whether Cpf1 crRNAs can also activate innate immune responses in human cells. We prepared a DNMT1-specific Acidaminococcus sp. Cpf1 (AsCpf1) crRNA via chemical synthesis or in vitro transcription using T7 polymerase (Fig. 3A). IFNB1, DDX58, and OAS2 mRNA levels were up-regulated by the 5′-ppp crRNA but not by the 5′-OH crRNA prepared by chemical synthesis or by treatment of the in vitro transcript with CIP (Fig. 3B–D). Activation of these IFN-mediated immune responses by the 5′-ppp crRNA was abrogated in DDX58 KO cells (Fig. 3E–G). Mutation frequencies associated with treatment with RNPs containing the 5′-ppp crRNA or the 5′-OH crRNA were comparable, showing that the cleavage activity of Cpf1 was not affected by the 5′-triphosphate group of the crRNA (Fig. 3H).
Figure 3.
RNA-sensing immune responses activated by the AsCpf1-associated crRNA. (A) Schematics of DNMT1-targeting crRNAs, which vary according to their preparation methods. crRNA sequences complementary to the DNMT1-target site are shown in bold. (B–D) Relative IFNB1 (B), DDX58 (C), and OAS2 (D) mRNA levels at 24 h after transfection. Error bars, SEM; n = 3. (E–G) Relative IFNB1 (E), DDX58 (F), and OAS2 (G) mRNA levels in WT and DDX58 KO HeLa cell lines at 24 h post-transfection. Error bars, SEM; n = 3. (H) Indel frequencies induced by the DNMT1-targeting AsCpf1 RNP were measured using next-generation sequencing (NGS). Statistical significances were calculated by t-test. (n.s.) Not significant, (Syn) chemically synthesized crRNA, (IVT) in vitro–transcribed crRNA, (± CIP) in vitro–transcribed crRNA with or without CIP treatment.
Cytotoxicity induced by 5′-ppp sgRNAs in primary human T cells
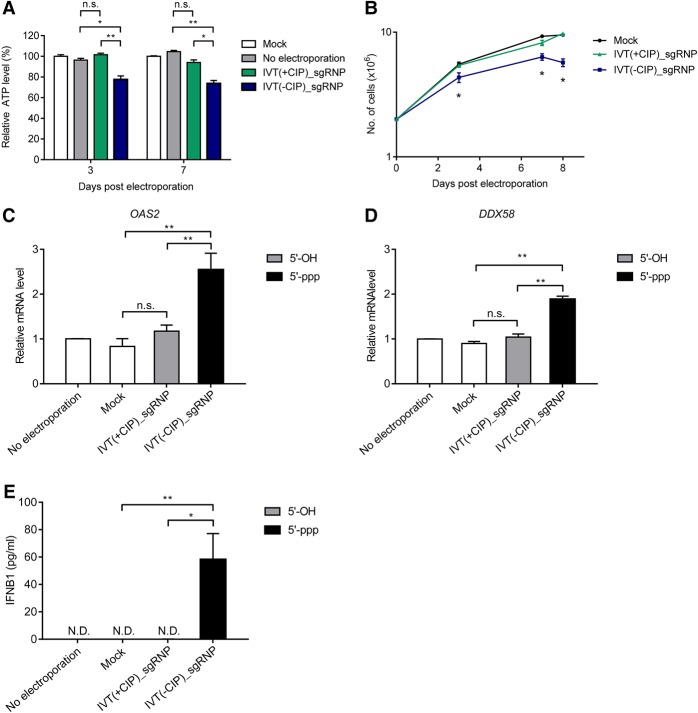
Genome editing in primary human T cells has been tested in clinics for treating cancer and human immunodeficiency virus type 1 (HIV-1) infection. We investigated whether 5′-triphosphate sgRNAs trigger innate immune responses in primary human T cells, causing cytotoxicity. We isolated primary CD4+ T cells from healthy volunteers (n = 3) and transfected a Cas9 RNP specific to the CCR5 gene, which encodes an essential coreceptor of HIV-1, into these cells via electroporation. We measured adenosine triphosphate (ATP) levels to monitor cell viability and found that the 5′-ppp gRNA treatment significantly reduced cell viability (*P < 0.05, **P < 0.01), whereas 5′-OH-sgRNA did not cause cytotoxicity (Fig. 4A). Furthermore, 5′-ppp gRNA-treated cells failed to divide after 7 d post-electroporation, whereas cells transfected with CIP-treated 5′-OH sgRNA in complex with the Cas9 protein were successfully expanded (Fig. 4B).
Figure 4.
5′-triphosphate induces RNA-sensing immune responses in human CD4+ T cells. A CCR5-targeting Cas9 RNP was delivered into primary CD4+ T cells using a MaxCyte electroporation system. (A) Cell viabilities at 3 and 7 d post-electroporation were determined by relative ATP level. (B) Live cell numbers from day 0 to 8 were counted after RNP delivery. Relative OAS2 (C) and DDX58 (D) mRNA levels were determined by real-time PCR 3 d post-electroporation. (E) IFNB1 protein levels in the supernatant were measured by ELISA. Error bars, SEM; n = 3. Statistical significances were calculated by t-test. (*) P < 0.05, (**) P < 0.01, (n.s.) not significant.
We next investigated whether the cytotoxicity of 5′-ppp sgRNA in primary CD4+ T cells was caused by RNA-sensing innate immune responses. As expected, OAS2 and DDX58 mRNA levels were up-regulated by the 5′-triphophate sgRNA (**P < 0.01) (Fig. 4C,D). The IFNB1 protein was also detected at day 3 post-electroporation (Fig. 4E). The 5′-OH sgRNA did not induce any detectable immune responses, compared to the mock control.
Highly efficient CCR5 knockout in human T cells using 5′-OH sgRNA
We next measured mutation frequencies in primary T cells using targeted deep sequencing. Removal of the 5′-triphosphate moiety from the sgRNA enhanced mutation efficiencies at the CCR5 target site in T cells. Thus, the 5′-OH sgRNA induced indels with a frequency of 94 ± 0.4% and 92 ± 0.8% at day 3 and 7 post-electroporation, respectively, whereas the 5′-ppp sgRNA inducled indels with a frequency of 87 ± 1% and 80 ± 1% (**P < 0.01) (Fig. 5A). Importantly, 5′-OH sgRNA-treated cells remained mutated at the CCR5 locus with a frequency of 95 ± 1% at day 25 post-treatment (Fig. 5B).
Figure 5.
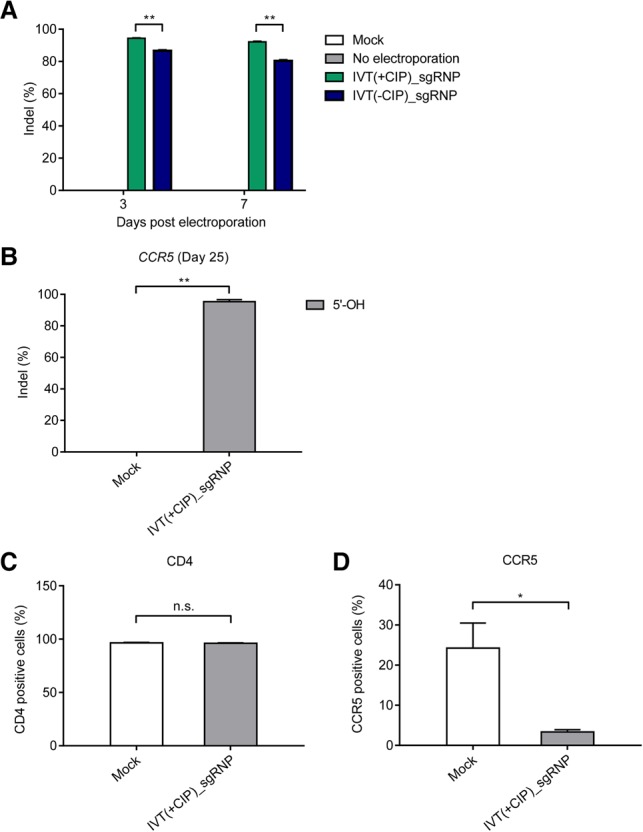
Removing the 5′-triphosphate moiety from gRNA improves CCR5 editing efficiency in primary human CD4+ T cells. (A) Indel frequencies at the CCR5 locus were calculated by targeted deep sequencing on day 3 and day 7 post-treatment. (B) Indel frequencies at the CCR5 locus were calculated by targeted deep sequencing on day 25 post-treatment. The percentages of cells positive for surface CD4 (C) and CCR5 protein (D) were determined by flow cytometry. Error bars, SEM; n = 3. Statistical significances were calculated by t-test. (*) P < 0.05, (**) P < 0.01, (n.s.) not significant.
To confirm the removal of the CCR5 protein from the surface of CD4+ T cells as a result of CRISPR gene editing, we performed flow cytometry. At day 7 post-electroporation, 96% of cells (both mock-treated and Cas9 RNP-treated) were CD4 positive (Fig. 5C; Supplemental Fig. S2). Among these CD4+ T cells, only 3.7% of cells treated with the Cas9 RNP were CCR5-positive, whereas 24.4% of mock-treated cells were CCR5-positive, showing an 85% reduction in the fraction of CCR5-positive cells upon Cas9 RNP treatment (Fig. 5D; Supplemental Fig. S2).
Discussion
The delivery of CRISPR RNP complexes that consist of Cas9 or Cpf1 protein and guide RNA into mammalian (Kim et al. 2014, 2016; Hur et al. 2016) and plant cells (Woo et al. 2015) and animal (Cho et al. 2013b; Sung et al. 2014) and human embryos (Ma et al. 2017) has been demonstrated as a promising approach for highly efficient and precise genome editing. CRISPR RNPs have been successfully delivered into retinal pigment epithelium cells (Kim et al. 2017b), neuronal cells (Staahl et al. 2017), and hair cells (Zuris et al. 2015) in mice, illustrating the broad potential for clinical applications. To prepare gRNAs for RNP delivery, in vitro transcription using phage RNA polymerases, such as T7 and SP6 polymerases, is the most accessible method because it is easier and more affordable than chemical synthesis for obtaining large amounts of RNA. In particular, the yield of ∼100-nt sgRNAs from chemical synthesis is very low (Hendel et al. 2015), and thus preparation of sgRNAs in amounts sufficient for transfecting 107∼109 cells by this method can be cost-prohibitive. However, in vitro transcription using phage RNA polymerases generates a triphosphate moiety at the 5′ end of the gRNA, and the resulting 5′-triphosphate single-stranded RNA (ssRNA) activates type I interferon-mediated immune responses in cells. Cytosolic ssRNAs bearing 5′-triphosphates are recognized by the DDX58 receptor (Pichlmair et al. 2006; Rehwinkel et al. 2010), but the preference of DDX58 for ssRNA ligands varies according to their concentration, sequence, length, conformation, and the level of shielding by proteins (Schlee and Hartmann 2016), and thus the minimal requirement of a biologically relevant ssRNA ligand for DDX58 activity has remained unclear. Here, we demonstrated that in vitro–transcribed gRNAs in CRISPR-Cas9 or Cpf1 RNPs indeed trigger RNA-sensing innate immune responses and result in cell death, which can be prevented by removal of the 5′-triphosphate group from the gRNA.
Human T cells can be gene-edited using CRISPR systems or other programmable nucleases for the treatment of various diseases, including cancer and acquired immune deficiency syndrome (AIDS). For example, a naturally occurring deletion in the CCR5 gene encoding an essential coreceptor of HIV confers resistance to HIV infection, providing the basis of cell therapy against HIV infection (Hendel et al. 2015; Hultquist et al. 2016; Xu et al. 2017). Here, we showed that delivery of RNP complexes containing gRNAs lacking the 5′-triphosphate rescued human CD4+ primary T cells from RNA-sensing innate immune responses, leading to efficient disruption of the CCR5 gene. Application of this strategy offers great promise for targeted cell therapy using CRISPR-Cas9, Cpf1, and Base Editor RNP complexes.
Methods
Preparation of recombinant Cas9 protein, Cpf1 protein, and guide RNA
Recombinant Cas9 protein was purchased from ToolGen. Recombinant Cpf1 protein was prepared as described previously (Hur et al. 2016). Cas9 crRNAs, tracrRNAs, and sgRNAs and Cpf1 crRNAs were synthesized by in vitro transcription using T7 RNA polymerase and a template oligonucleotide (Supplemental Table S1) as described previously (Kim et al. 2014, 2016). CIP (New England BioLabs) was used to remove the 5′-triphosphate from gRNAs as follows: 10 µg of in vitro–transcribed RNA was treated with 250 units of CIP for 3 h at 37°C in the presence of 100 units of RNase inhibitor (New England BioLabs). Following CIP treatment, the RNA was cleaned up using a miRNeasy Mini kit (Qiagen). Chemically synthesized RNAs, which were purified using high-performance liquid chromatography (HPLC), were purchased from Integrated DNA Technologies (IDT).
Cell culture and transfection
HeLa (ATCC, CCL-2) cells were maintained in Dulbecco's Modified Eagle's Medium (DMEM) supplemented with 100 units/mL penicillin, 100 µg/mL streptomycin, 0.1 mM nonessential amino acids, and 10% fetal bovine serum (FBS). Mouse embryonic fibroblast cells were grown in DMEM with 20% fetal bovine serum. HeLa cells and MEF cells were transfected with 100 nM of purified Cas9 protein or Cpf1 protein and 300 nM of gRNA using Lipofectamine 2000 reagent (Invitrogen), as described in Zuris et al. (2015).
Isolation of primary human CD4+ T cells
Peripheral blood was collected from healthy donors at the National Medical Center of Korea after informed consent with Institutional Review Board approval. Peripheral blood mononuclear cells (PBMCs) were isolated by density gradient centrifugation using Ficoll-Paque solution (GE Healthcare). Using a magnetic activated cell sorting (MACS) CD4+ T cell isolation kit (Miltenyi Biotec), CD4+ T cells were purified by negative selection. Isolated CD4+ T cells were cultured in RPMI 1640 medium supplemented with 2 mM glutamine, 1% (v/v) nonessential amino acids, 1% (v/v) sodium pyruvate, 1% (v/v) HEPES, penicillin (50 U/mL), streptomycin (50 µg/mL), and 10% (v/v) FBS (medium and culture supplements from Invitrogen). Within 2 h of isolation, CD4+ T cells were activated with CD3/CD28 Dynabeads (Thermo Fisher Scientific). The CD3/CD28 beads were magnetically removed 72 h after activation, before delivery of RNPs for CCR5 knockout. For long-term expansion of CD4+ T cells, CD3/CD28 Dynabeads were added at 1:1 ratio (cell to bead) again at day 8 post-electroporation (day 11 post-initial activation) (Levine et al. 1997).
RNP delivery into primary human CD4+ T cells
RNPs were delivered into T cells via electroporation. Bead-free activated CD4+ T cells were mixed with purified Cas9 protein and sgRNA complexes in MaxCyte buffer and transferred into an OC-100 processing assembly, followed by electroporation using a MaxCyte-STX transfection system (MaxCyte Systems) according to the manufacturer's instructions. After electroporation, cells were seeded on 96-well plates with 20 units/mL of recombinant human IL2.
Targeted deep sequencing for mutation detection
Genomic DNA was harvested with a DNeasy Blood & Tissue kit (Qiagen) after RNP transfection. The target regions were PCR-amplified using appropriate primer sets (Supplemental Table S1) for next-generation sequencing library construction. TruSeq HT Dual Index primers were used for labeling each sample. The resulting libraries were subjected to paired-end read sequencing using MiSeq (Illumina).
Measurement of relative mRNA levels by quantitative PCR
Transfected cells were lysed and mRNA was purified either 24 h or 3 d after transfection using a NucleoSpin RNA kit (MACHEREY-NAGEL). cDNAs were synthesized by reverse transcription using a miScript II RT kit (Qiagen) and quantified by qPCR using the primers listed in Supplemental Table S1. The qPCR in primary human CD4+ T cells was performed using TaqMan gene expression assays (Thermo Fisher Scientific) with the following primers: DDX58 (Hs01061436_m1), OAS2 (Hs00942643_m1), 18SrRNA (Hs03928990_g1).
ELISA
The amount of IFNB1 secreted into the growth medium was determined using a Human IFN beta ELISA kit (PBL Assay Science). The medium from RNP-transfected cells was collected at either 24 h or 3 d post-transfection. The IFNB1 concentration was measured according to the manufacturer's protocol.
Cell viability measurements
In the case of HeLa cells, 5 × 103 of either wild-type or DDX58 KO cells were plated in 96-well plates 1 d before RNP transfection. At day 3 after transfection, cell viability was determined by a WST-1 assay (Roche) according to the manufacturer's protocol. In the case of primary CD4+ T cells, they were plated in 96-well plates after electroporation to deliver RNPs. At day 3 and 7 after electroporation, the ATP level in the supernatants, a measure of cell viability, was determined using the CellTiter-Glo substrate (Promega).
Data access
The deep sequencing data from this study have been submitted to the NCBI Sequence Read Archive (SRA; https://www.ncbi.nlm.nih.gov/sra) under accession numbers SRP121089 and SRP129873.
Supplementary Material
Acknowledgments
This work was supported by a grant from the Institute for Basic Science (IBS-R021-D1).
Footnotes
[Supplemental material is available for this article.]
Article published online before print. Article, supplemental material, and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.231936.117.
Freely available online through the Genome Research Open Access option.
References
- Cho SW, Kim S, Kim JM, Kim JS. 2013a. Targeted genome engineering in human cells with the Cas9 RNA-guided endonuclease. Nat Biotechnol 31: 230–232. [DOI] [PubMed] [Google Scholar]
- Cho SW, Lee J, Carroll D, Kim JS, Lee J. 2013b. Heritable gene knockout in Caenorhabditis elegans by direct injection of Cas9–sgRNA ribonucleoproteins. Genetics 195: 1177–1180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deltcheva E, Chylinski K, Sharma CM, Gonzales K, Chao Y, Pirzada ZA, Eckert MR, Vogel J, Charpentier E. 2011. CRISPR RNA maturation by trans-encoded small RNA and host factor RNase III. Nature 471: 602–607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dever DP, Bak RO, Reinisch A, Camarena J, Washington G, Nicolas CE, Pavel-Dinu M, Saxena N, Wilkens AB, Mantri S, et al. 2016. CRISPR/Cas9 β-globin gene targeting in human haematopoietic stem cells. Nature 539: 384–389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendel A, Bak RO, Clark JT, Kennedy AB, Ryan DE, Roy S, Steinfeld I, Lunstad BD, Kaiser RJ, Wilkens AB, et al. 2015. Chemically modified guide RNAs enhance CRISPR-Cas genome editing in human primary cells. Nat Biotechnol 33: 985–989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hultquist JF, Schumann K, Woo JM, Manganaro L, McGregor MJ, Doudna J, Simon V, Krogan NJ, Marson A. 2016. A Cas9 ribonucleoprotein platform for functional genetic studies of HIV-host interactions in primary human T cells. Cell Rep 17: 1438–1452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hur JK, Kim K, Been KW, Baek G, Ye S, Hur JW, Ryu SM, Lee YS, Kim JS. 2016. Targeted mutagenesis in mice by electroporation of Cpf1 ribonucleoproteins. Nat Biotechnol 34: 807–808. [DOI] [PubMed] [Google Scholar]
- Jinek M, Chylinski K, Fonfara I, Hauer M, Doudna JA, Charpentier E. 2012. A programmable dual-RNA–guided DNA endonuclease in adaptive bacterial immunity. Science 337: 816–821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H, Kim JS. 2014. A guide to genome engineering with programmable nucleases. Nat Rev Genet 15: 321–334. [DOI] [PubMed] [Google Scholar]
- Kim DH, Longo M, Han Y, Lundberg P, Cantin E, Rossi JJ. 2004. Interferon induction by siRNAs and ssRNAs synthesized by phage polymerase. Nat Biotechnol 22: 321–325. [DOI] [PubMed] [Google Scholar]
- Kim S, Kim D, Cho SW, Kim J, Kim JS. 2014. Highly efficient RNA-guided genome editing in human cells via delivery of purified Cas9 ribonucleoproteins. Genome Res 24: 1012–1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D, Kim J, Hur JK, Been KW, Yoon SH, Kim JS. 2016. Genome-wide analysis reveals specificities of Cpf1 endonucleases in human cells. Nat Biotechnol 34: 863–868. [DOI] [PubMed] [Google Scholar]
- Kim H, Kim ST, Ryu J, Kang BC, Kim JS, Kim SG. 2017a. CRISPR/Cpf1-mediated DNA-free plant genome editing. Nat Commun 8: 14406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim K, Park SW, Kim JH, Lee SH, Kim D, Koo T, Kim KE, Kim JH, Kim JS. 2017b. Genome surgery using Cas9 ribonucleoproteins for the treatment of age-related macular degeneration. Genome Res 27: 419–426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim K, Ryu SM, Kim ST, Baek G, Kim D, Lim K, Chung E, Kim S, Kim JS. 2017c. Highly efficient RNA-guided base editing in mouse embryos. Nat Biotechnol 35: 435–437. [DOI] [PubMed] [Google Scholar]
- Levine BL, Bernstein WB, Connors M, Craighead N, Lindsten T, Thompson CB, June CH. 1997. Effects of CD28 costimulation on long-term proliferation of CD4+ T cells in the absence of exogenous feeder cells. J Immunol 159: 5921–5930. [PubMed] [Google Scholar]
- Liang Z, Chen K, Li T, Zhang Y, Wang Y, Zhao Q, Liu J, Zhang H, Liu C, Ran Y, et al. 2017. Efficient DNA-free genome editing of bread wheat using CRISPR/Cas9 ribonucleoprotein complexes. Nat Commun 8: 14261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma H, Marti-Gutierrez N, Park SW, Wu J, Lee Y, Suzuki K, Koski A, Ji D, Hayama T, Ahmed R, et al. 2017. Correction of a pathogenic gene mutation in human embryos. Nature 548: 413–419. [DOI] [PubMed] [Google Scholar]
- Pichlmair A, Schulz O, Tan CP, Naslund TI, Liljestrom P, Weber F, Reis e Sousa C. 2006. RIG-I-mediated antiviral responses to single-stranded RNA bearing 5′-phosphates. Science 314: 997–1001. [DOI] [PubMed] [Google Scholar]
- Ramakrishna S, Kwaku Dad AB, Beloor J, Gopalappa R, Lee SK, Kim H. 2014. Gene disruption by cell-penetrating peptide-mediated delivery of Cas9 protein and guide RNA. Genome Res 24: 1020–1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rehwinkel J, Tan CP, Goubau D, Schulz O, Pichlmair A, Bier K, Robb N, Vreede F, Barclay W, Fodor E, et al. 2010. RIG-I detects viral genomic RNA during negative-strand RNA virus infection. Cell 140: 397–408. [DOI] [PubMed] [Google Scholar]
- Schlee M, Hartmann G. 2016. Discriminating self from non-self in nucleic acid sensing. Nat Rev Immunol 16: 566–580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staahl BT, Benekareddy M, Coulon-Bainier C, Banfal AA, Floor SN, Sabo JK, Urnes C, Munares GA, Ghosh A, Doudna JA. 2017. Efficient genome editing in the mouse brain by local delivery of engineered Cas9 ribonucleoprotein complexes. Nat Biotechnol 35: 431–434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun L, Wu J, Du F, Chen X, Chen ZJ. 2013. Cyclic GMP-AMP synthase is a cytosolic DNA sensor that activates the type I interferon pathway. Science 339: 786–791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sung YH, Kim JM, Kim HT, Lee J, Jeon J, Jin Y, Choi JH, Ban YH, Ha SJ, Kim CH, et al. 2014. Highly efficient gene knockout in mice and zebrafish with RNA-guided endonucleases. Genome Res 24: 125–131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo JW, Kim J, Kwon SI, Corvalan C, Cho SW, Kim H, Kim SG, Kim ST, Choe S, Kim JS. 2015. DNA-free genome editing in plants with preassembled CRISPR-Cas9 ribonucleoproteins. Nat Biotechnol 33: 1162–1164. [DOI] [PubMed] [Google Scholar]
- Xu L, Yang H, Gao Y, Chen Z, Xie L, Liu Y, Liu Y, Wang X, Li H, Lai W, et al. 2017. CRISPR/Cas9-mediated CCR5 ablation in human hematopoietic stem/progenitor cells confers HIV-1 resistance in vivo. Mol Ther 25: 1782–1789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zetsche B, Gootenberg JS, Abudayyeh OO, Slaymaker IM, Makarova KS, Essletzbichler P, Volz SE, Joung J, van der Oost J, Regev A, et al. 2015. Cpf1 is a single RNA-guided endonuclease of a class 2 CRISPR-Cas system. Cell 163: 759–771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuris JA, Thompson DB, Shu Y, Guilinger JP, Bessen JL, Hu JH, Maeder ML, Joung JK, Chen ZY, Liu DR. 2015. Cationic lipid-mediated delivery of proteins enables efficient protein-based genome editing in vitro and in vivo. Nat Biotechnol 33: 73–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.