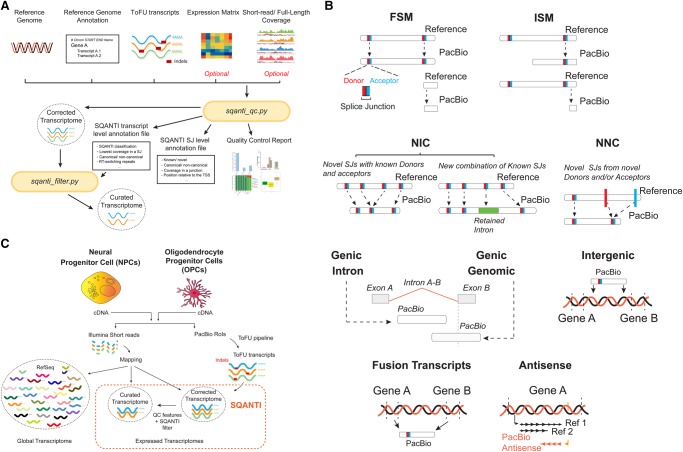
Figure 1.
Overview of the experimental model and SQANTI analysis. (A) SQANTI workflow. Two main functions are part of SQANTI. sqanti_qc.py uses as input files a FASTA file with transcript sequences, the reference genome in FASTA format, a GTF annotation file, and optionally, full-length and short-read expression files. The function returns a reference-corrected transcriptome, transcript-level and junction-level files with structural and quality descriptors, and a QC graphical report. sqanti_filter.py takes the reference-corrected transcriptome and the transcript-level descriptors file to return a curated transcriptome from which artifacts have been removed. (B) SQANTI classification of transcripts according to their splice junctions and donor and acceptor sites. Splice donors and acceptors are indicated in red and blue, respectively. (SJ) splice junction, (FSM) Full Splice Match, (ISM) Incomplete Splice Match, (NIC) Novel in Catalog, (NNC) Novel Not in Catalog. (C) Experimental system and data processing pipeline. RNA isolated from neural progenitor cells (NPCs) and oligodendrocyte precursor cells (OPCs) was retrotranscribed separately into cDNA and sequenced both by long-read PacBio and short-read Illumina technologies. All PacBio RoIs were joined and processed by the ToFU pipeline to obtain consensus transcripts. Residual (indel) errors were eliminated by comparison to the reference genome to generate a corrected transcriptome, and false transcripts were removed using a SQANTI filter to result in a curated transcriptome. Illumina short reads were mapped against the RefSeq murine transcriptome annotation, the corrected, and the curated PacBio transcriptomes.