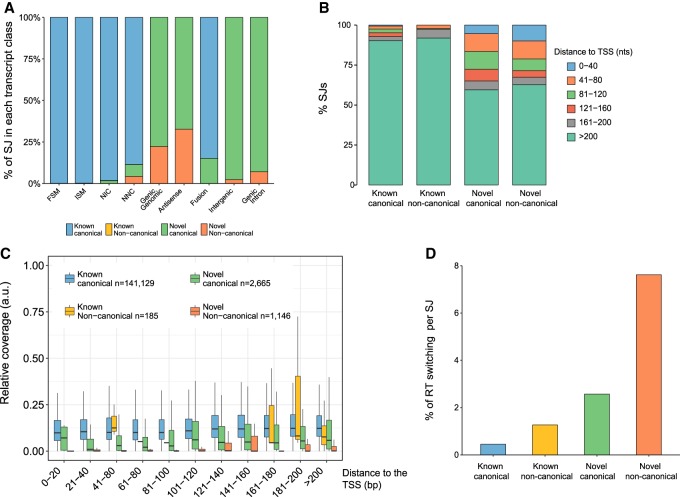
Figure 3.
Splice junction characterization in the corrected PacBio transcriptome. (A) Distribution of splice junction (SJ) types across SQANTI categories. NNC, Genic Genomic, Antisense, Intergenic, and Genic Intron are enriched in noncanonical SJs. n = 76,757 SJ for FSM, n = 13,802 for ISM, n = 27,368 for NIC, n = 26,509 for NNC, n = 51 for Genic Genomic, n = 49 for Antisense, n = 494 for Fusion, n = 86 for Intergenic, and n = 55 for Genic Intron. (B) Distribution of the SJs according to their distance to the transcription start site. (C) Relative coverage by short reads of SJs as a function of their class and distance to the TSS. (a.u.) Arbitrary units. (D) Detection of RT switching direct repetitions by SQANTI algorithm across SJ types.