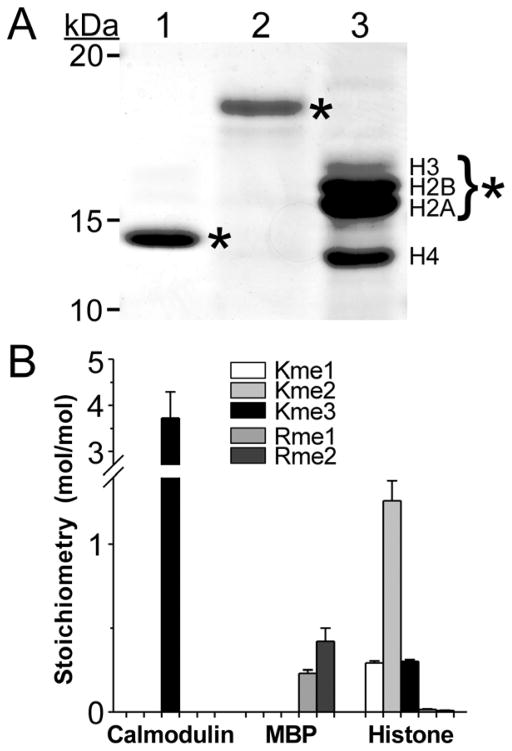
Fig. 3.
Quantification of methylation stoichiometry in protein standards. (A) Coomassie-blue stained SDS-PAGE gel of input protein standards calmodulin (lane 1, 1 μg), MBP (lane 2, 2 μg) and mixed histones (lane 3, 3 μg). Bands excised for hydrolysis and LC-MS/MS analysis are marked by asterisks. The excised histone bands consisted of core histones H2A, H2B and H3. (B) Methylation stoichiometry determined by LC-MS/MS and eqns. 1–4. Bar height indicates contributions of 1meK, 2meK, 3meK, 1meR and 2meR to mol methyl/mol protein in each sample (triplicate determination) whereas error bars represent the standard deviation.