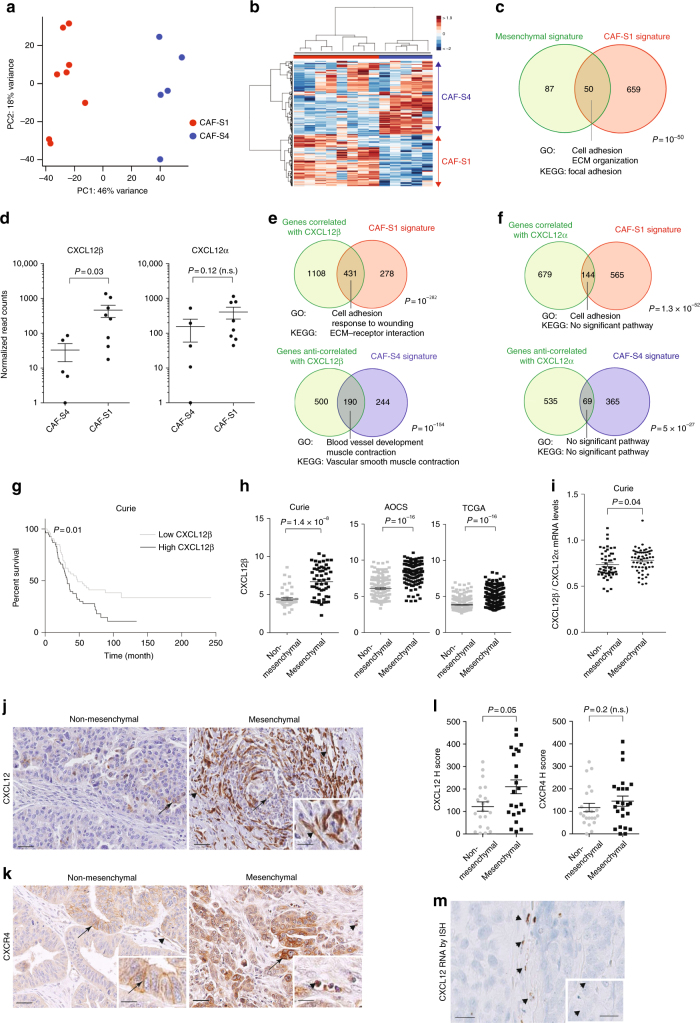
Fig. 4.
CXCL12β discriminates CAF-S1 and CAF-S4 cells. a PCA based on the 500 most variant transcripts differentiating CAF-S1 (red) and CAF-S4 (blue). b HC (500 most variant transcripts) using Ward’s method with Euclidean distances. Each column represents a CAF subset and each row a gene. Color saturation shows gene expression deviation from the mean (above in red, below in blue). c Venn diagram showing overlap between mesenchymal signature (defined in ref. 7) and CAF-S1 signature (Supplementary Data 1). P value is from hypergeometric test. d Scatter plots of CXCL12α (NM_000609) or CXCL12β (NM_199168) mRNA levels in CAF-S1 and CAF-S4 subsets. e, f Venn diagrams showing overlap between genes correlated or anti-correlated with CXCL12β (e) or CXCL12α (f) and CAF-S1 or CAF-S4 signatures (Supplementary Data 1 and 2). P values are from hypergeometric test. g Kaplan−Meier curves of overall survival according to low- and high-CXCL12β mRNA levels (N = 53 in low-CXCL12β subgroup and N = 54 patients in high-CXCL12β subgroup, Institut Curie). P value is based on log-rank test. h Scatter plots showing CXCL12β mRNA levels in mesenchymal and non-mesenchymal HGSOC from the Institut Curie, AOCS, and TCGA cohorts. Data (log2 of probeset (203666_at) intensity) are shown as mean ± SEM. P values are from Mann-Whitney test. i Scatter plot showing ratio of CXCL12α and CXCL12β expression levels in mesenchymal and non-mesenchymal HGSOC of the Institut Curie cohort. Data are shown as mean ± SEM. P values are from Mann-Whitney test. j, k Representative views of CXCL12 (j) and CXCR4 (k) immunostaining in HGSOC. Scale bar, 50 μm (low magnification) and 20 μm (inset). l Scatter plots showing histological scores (H score) of CXCL12 and CXCR4 proteins. H score corresponds to the percentage of positive cells (in CAF and at epithelial cell surface, arrows in j, k) multiplied by the staining intensity. Data are shown as mean ± SEM. P values are from Mann-Whitney test. m Representative view of CXCL12 mRNA detected in fibroblasts by in situ hybridization, using RNAscope® Technology on HGSOC tissue section. Scale bar, 20 μm (low magnification) and 6 μm (inset)