Fig. 2.
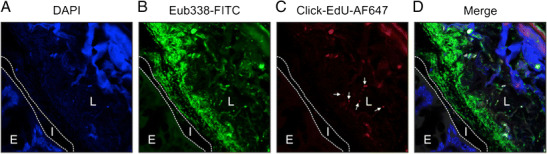
In vivo tracking and improved spatial resolution of gut-microbiome interactions. A mouse colon was fixed in Carnoy’s solution to preserve mucus layer architecture. a The section was counterstained with 4′,6-diamidino-2-phenylindole (DAPI) indicating the colonic epithelium (E) in the left lower corner, the interlaced (I) mucus layer (indicated by the two dotted lines), which is devoid of bacteria, and the colonic lumen (L) on the right side. The structure in the upper right depicts a plant component. b Bacteria were stained by FISH using a fluorescein isothiocyanate (FITC) pan-specific EUB338 probe (green) which covers approximately 90% of the domain bacteria. c In this experiment, mice were gavaged with the gut bacterium Alistipes finegoldii (phylum Bacteroidetes), which were cultured in the presence of the thymidine analog 5-ethynyl-2′-deoxyuridine (EdU). Metabolically active Alistipes was tracked utilizing click chemistry that is based on a copper-catalyzed covalent reaction between an alkyne (within the EdU) and an AlexaFluor® 647-containing azide. Note that the bacterium colonizes the luminal site of the colon, not the mucus layer (arrows). d The right picture shows the merged panels. Cells were imaged on a Zeiss Axioobserver Z1 microscope equipped with a LSM700 confocal unit. Original magnification 400×
