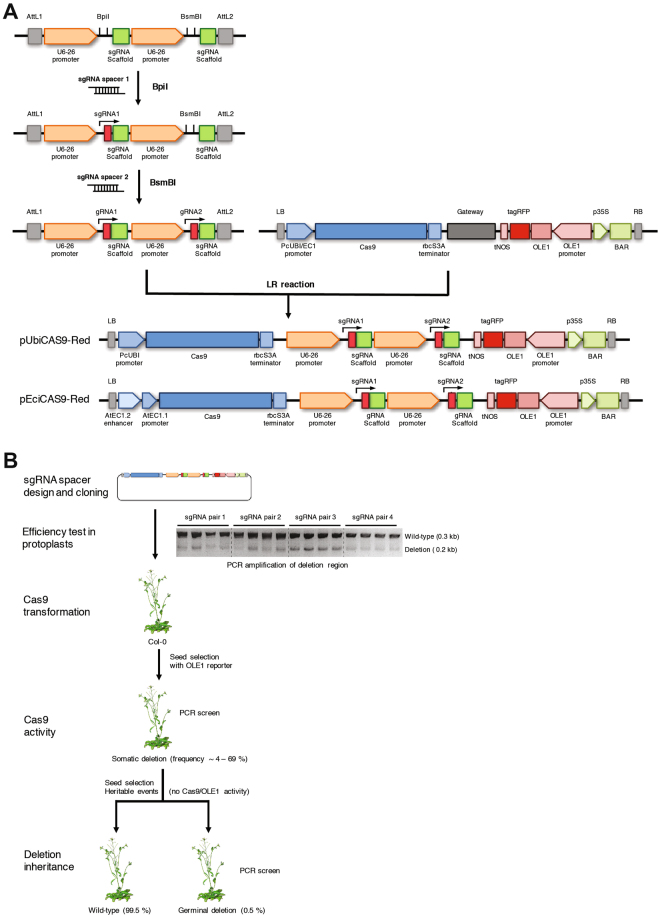
Figure 1.
Methodology for the generation of targeted chromosomal deletions in Arabidopsis. (A) Schematic assembly of the CRISPR/Cas9 vectors. Briefly, 24 nt DNA oligos containing the sequences of the two sgRNAs were used to generate a double-stranded DNA molecule and cloned into the entry vector with the two restriction enzyme sites BpiI and BsmBI, respectively. A LR reaction between the entry vector containing the sgRNAs and the binary vector generated the final Cas9 expression vector. (B) The efficiency of the Cas9 expression vector was determined by transfection of Arabidopsis protoplast. Selected vectors were integrated using floral dipping. Positive transformants were selected using a seed-specific fluorescent reporter. Cas9 nuclease activity in independent transgenic lines was confirmed by PCR using oligos flanking the deletion. Heritable deletion events were identified in plants lacking Cas9 activity after selection for seeds lacking the seed-specific fluorescent reporter. PCR was carried out using primers JD460 and JD461.